N.B.: This script is optimised for the open-source browser Mozilla Firefox v.123.0
Some functions might not work in other webbrowsers...
The map is not the territory.
The menu is not the meal.
(Cf. Korzybski's general semantics argument)
Dk default
# Enable this universe
options(repos = c(
ggseg = 'https://ggseg.r-universe.dev',
CRAN = 'https://cloud.r-project.org'))
# Install some packages
#install.packages('ggsegDefaultExtra')
##########################
library(ggsegDefaultExtra)
library(ggseg)
library(ggplot2)
plot(dkextra) +
theme(legend.position = "bottom",
legend.text = element_text(size = 7)) +
guides(fill = guide_legend(ncol = 3))
##########################
ggseg(atlas = dkextra, mapping = aes(fill = region)) +
scale_fill_brain("dk") +
theme(legend.position = "bottom",
legend.text = element_text(size = 7)) +
guides(fill = guide_legend(ncol = 3))
##########################
library(ggseg3d)
library(dplyr)
ggseg3d(atlas = hcpa_3d) %>%
add_glassbrain("left") %>%
pan_camera("right lateral")
##########################
ggseg3d(atlas = hcpa_3d) %>%
add_glassbrain("left") %>%
pan_camera("right lateral")
##########################
# Print all rows of dkextra
print(dkextra, n = Inf)
# Capture the output of print(dkextra)
output <- capture.output(print(dkextra, n = Inf))
# Open a text file for writing
file_path <- "dkextra.txt"
writeLines(output, file_path)
Based on: Mowinckel AM, Vidal-Piñeiro D. Visualization of Brain Statistics With R Packages ggseg and ggseg3d.
Advances in Methods and Practices in Psychological Science. 2020; 3(4):466-483. doi:10.1177/2515245920928009
Brodmann, K. Vergleichende Lokalisationslehre der Grosshirnrinde in ihren Prinzipien dargestellt auf Grund des Zellenbaues (J. A. Barth, 1909); Brodmann’s Localization in the Cerebral Cortex (Smith Gordon, 1994) [transl. Garey, L.J.]
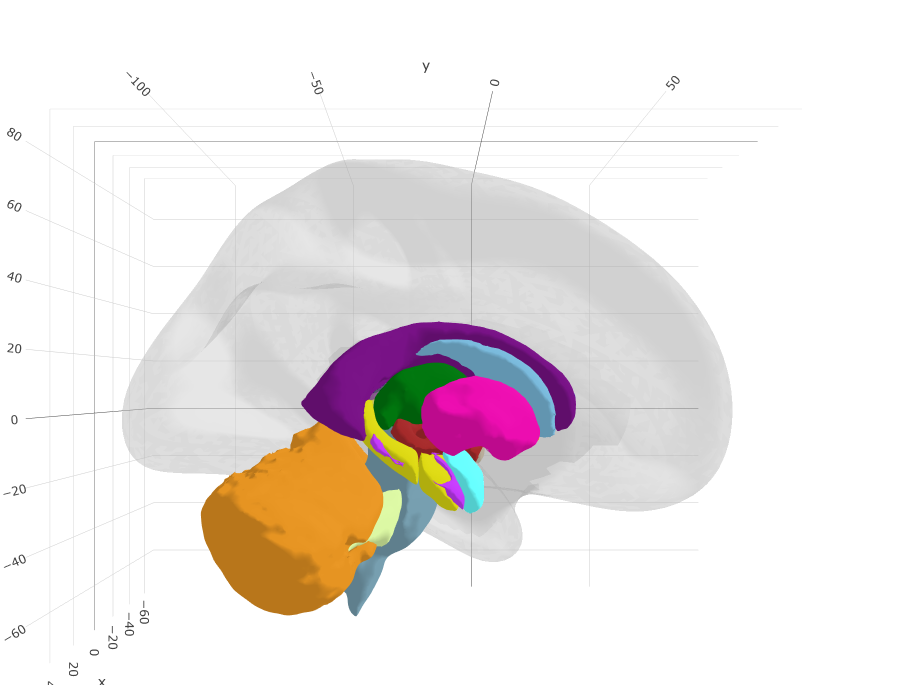
Tracula
Tracula (TRActs Constrained by UnderLying Anatomy)
URL: https://github.com/ggseg/ggsegTracula
This package contains a tractography dataset for plotting the 'Tracula' white matter tracts with ggseg and ggseg3d.
Based on: Yendiki et al. (2011) Automated probabilistic reconstruction of white-matter pathways in health and disease using an atlas of the underlying anatomy. Front. Neuroinform. 5:23. doi: 10.3389/fninf.2011.00023
Cf. Using diffusion MRI data acquired with ultra-high gradients to improve tractography in routine-quality data. Maffei C, Lee C, Planich M, Ramprasad M, Ravi N, Trainor D, Urban Z, Kim M, Jones R, Henin A, Hofmann S, Pizzagalli D, Auerbach R, Gabrieli J, Whitfield-Gabrieli S, Greve D, Haber N, Yendiki A (2021). NeuroImage 245:118706.
#See also https://surfer.nmr.mgh.harvard.edu/fswiki/Tracula
# Enable this universe
options(repos = c(
ggseg = 'https://ggseg.r-universe.dev',
CRAN = 'https://cloud.r-project.org'))
# Install some packages
install.packages('ggsegTracula')
# install.packages("remotes")
remotes::install_github("LCBC-UiO/ggsegTracula")
library(ggseg)
library(ggplot2)
library(ggseg3d)
library(ggsegTracula)
library(dplyr)
#>
#> Attaching package: 'dplyr'
#> The following objects are masked from 'package:stats':
#>
#> filter, lag
#> The following objects are masked from 'package:base':
#>
#> intersect, setdiff, setequal, union
plot(tracula) +
theme(legend.position = "bottom",
legend.text = element_text(size = 9)) +
guides(fill = guide_legend(ncol = 3))
ggseg3d(atlas = tracula_3d) %>%
add_glassbrain() %>%
pan_camera("right lateral")
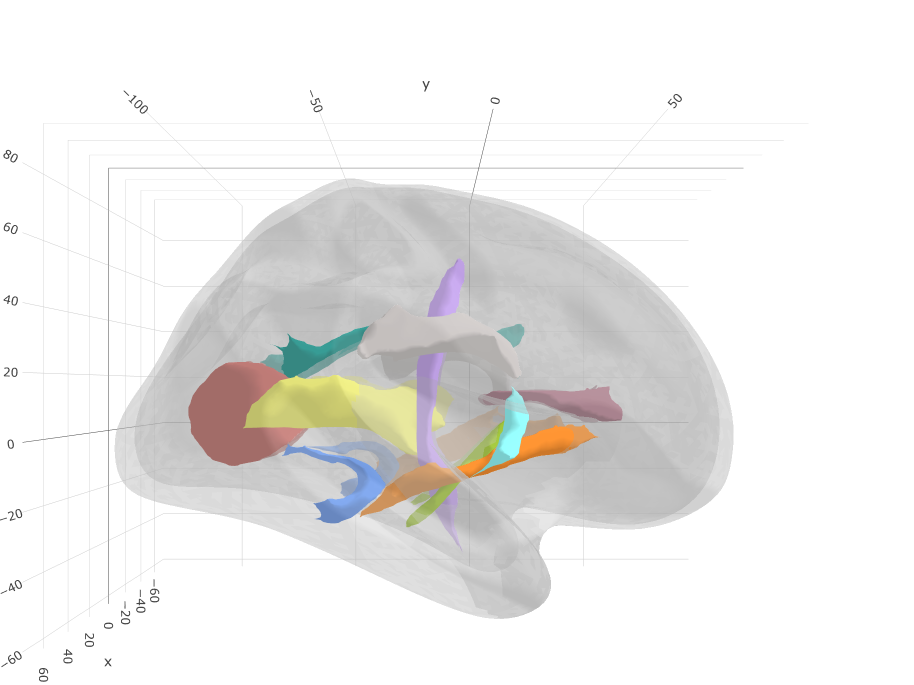
Subcortical tracts
Based on: Hua, K., Zhang, J., Wakana, S., Jiang, H., Li, X., Reich, D. S., Calabresi, P. A., Pekar, J. J., van Zijl, P. C., & Mori, S. (2008). Tract probability maps in stereotaxic spaces: analyses of white matter anatomy and tract-specific quantification. NeuroImage, 39(1), 336–347. https://doi.org/10.1016/j.neuroimage.2007.07.053
# Enable this universe
options(repos = c(
ggseg = 'https://ggseg.r-universe.dev',
CRAN = 'https://cloud.r-project.org'))
# Install some packages
install.packages('ggsegJHU')
library(ggsegJHU)
library(ggseg)
library(ggplot2)
plot(jhu) +
theme(legend.position = "bottom",
legend.text = element_text(size = 7)) +
guides(fill = guide_legend(ncol = 2))
library(ggseg3d)
library(dplyr)
ggseg3d(atlas = jhu_3d) %>%
add_glassbrain("left") %>%
pan_camera("right lateral")
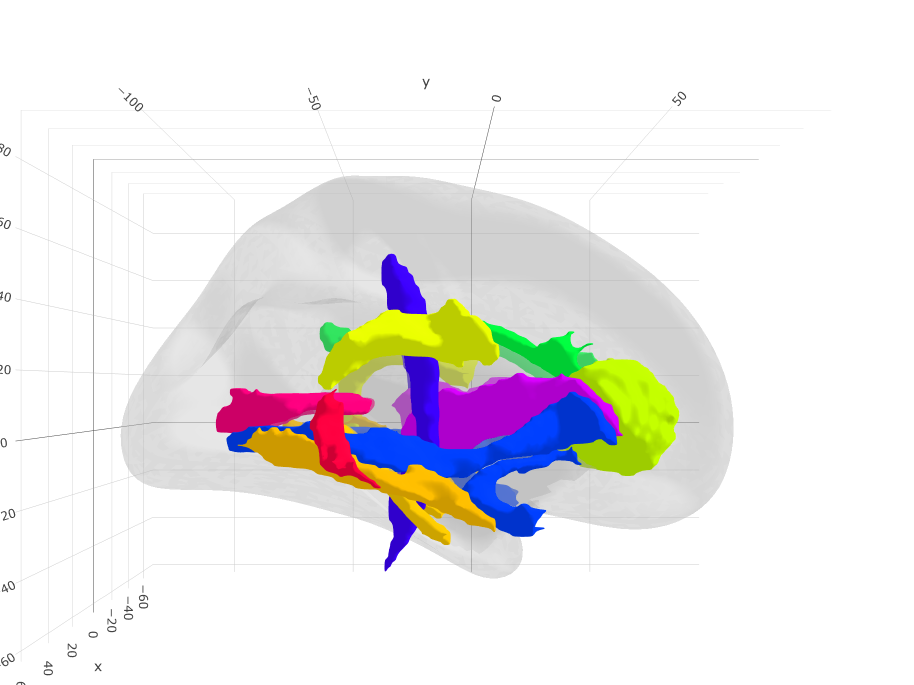
Aicha
Based on: Joliot, M., Jobard, G., Naveau, M., Delcroix, N., Petit, L., Zago, L., … & Tzourio-Mazoyer, N. (2015). AICHA: An atlas of intrinsic connectivity of homotopic areas. Journal of neuroscience methods, 254, 46-59.
# Enable this universe
options(repos = c(
ggseg = 'https://ggseg.r-universe.dev',
CRAN = 'https://cloud.r-project.org'))
#####################
library(ggseg)
library(ggseg3d)
library(ggsegAicha)
library(ggplot2)
plot(aicha) +
theme(legend.position = "bottom",
legend.text = element_text(size = 9)) +
guides(fill = guide_legend(ncol = 6))
######################
library(dplyr)
ggseg3d(atlas = aicha_3d) %>%
add_glassbrain() %>%
pan_camera("right lateral")
Attention networks
Based on: Yeo, B. T., Krienen, F. M., Sepulcre, J., Sabuncu, M. R., Lashkari, D., Hollinshead, M., Roffman, J. L., Smoller, J. W., Zöllei, L., Polimeni, J. R., Fischl, B., Liu, H., & Buckner, R. L. (2011). The organization of the human cerebral cortex estimated by intrinsic functional connectivity. Journal of neurophysiology, 106(3), 1125–1165. https://doi.org/10.1152/jn.00338.2011
#See also: https://surfer.nmr.mgh.harvard.edu/fswiki/CorticalParcellation_Yeo2011
# Enable this universe
options(repos = c(
ggseg = 'https://ggseg.r-universe.dev',
CRAN = 'https://cloud.r-project.org'))
# Install some packages
install.packages('ggsegYeo2011')
library(ggsegYeo2011)
library(ggplot2)
library(ggseg)
plot(yeo7) +
theme(legend.position = "bottom",
legend.text = element_text(size = 9)) +
guides(fill = guide_legend(ncol = 3))
##################
plot(yeo17) +
theme(legend.position = "bottom",
legend.text = element_text(size = 9)) +
guides(fill = guide_legend(ncol = 3))
##############
library(ggseg3d)
library(dplyr)
ggseg3d(atlas = yeo7_3d) %>%
pan_camera("right lateral")
#############
ggseg3d(atlas = yeo17_3d) %>%
pan_camera("right lateral")
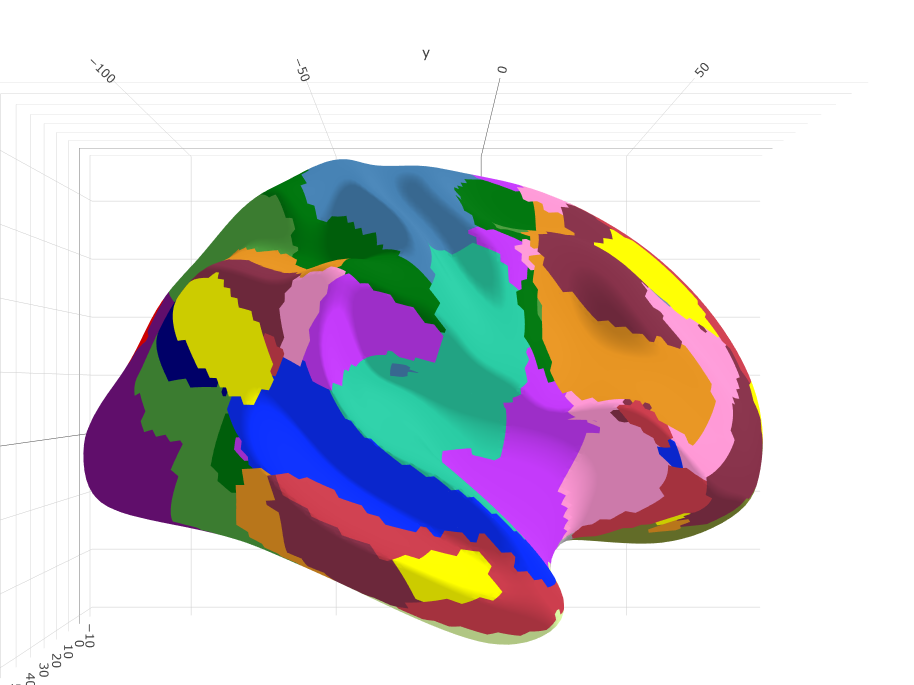
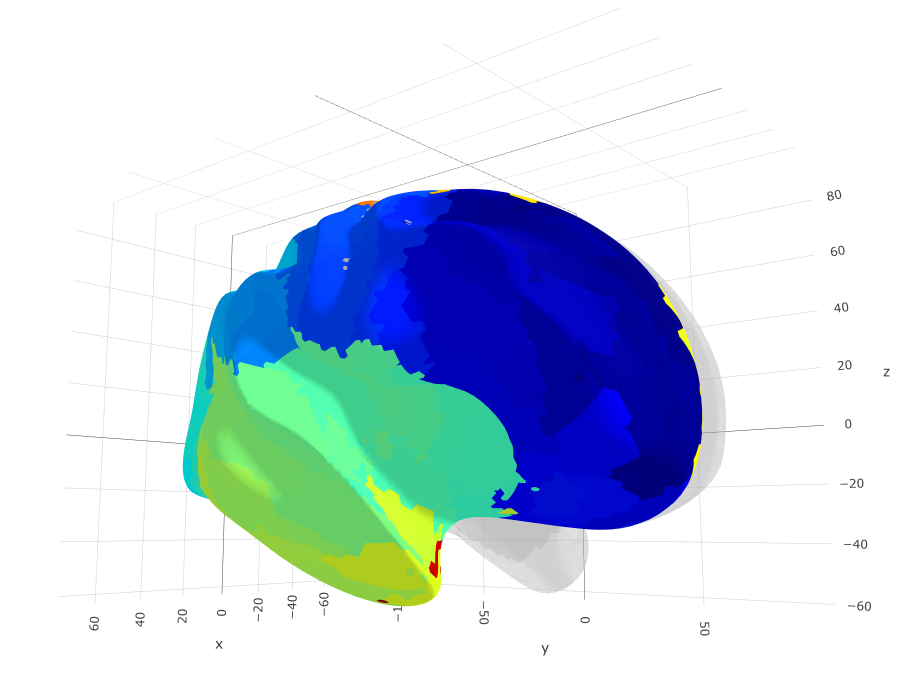
Glasser's parcellation for Human Connectome Project (HCP)
# Enable this universe
options(repos = c(
ggseg = 'https://ggseg.r-universe.dev',
CRAN = 'https://cloud.r-project.org'))
# Install some packages
install.packages('ggsegGlasser')
library(ggsegGlasser)
library(ggseg)
#> Warning: package 'ggseg' was built under R version 4.1.1
#> Loading required package: ggplot2
library(ggplot2)
plot(glasser) +
theme(legend.position = "bottom",
legend.text = element_text(size = 9)) +
guides(fill = guide_legend(ncol = 7))
#####################
library(ggseg3d)
library(dplyr)
ggseg3d(atlas = glasser_3d) %>%
pan_camera("right lateral")
Ryōkan (大愚)
Relying upon a finger, we see the moon
Relying upon the moon, we understand the finger.
— • —
Moon and finger
are neither the same nor different.
Having seen reality as it is,
There is neither moon nor finger.
— • —
Śuraṅgama Sūtra:
"You and others like you still listen to the Dharma with conditioned minds, and therefore you fail to understand its real nature. Consider this example: Suppose someone is pointing to the moon to show it to another person. That other person, guided by the pointing finger, should now look at the moon. But if he looks instead at the finger, taking it to be the moon, not only does he fail to see the moon, but he is mistaken, too, about the finger. He has confused the finger, with which someone is pointing to the moon, with the moon, which is being pointed to."
* * *
Hsüan Hua and Buddhist Text Translation Society, eds., The Śuraṅgama Sutra: A New Translation (Ukiah, Calif: Buddhist Text Translation Society, 2009), “Visual Awareness Is Not Dependent upon Conditions.”

ggseg
34 repositories, 32 followers.
icon-download Download ggseg-main.zip
“Contains ggplot2 geom for plotting brain atlases using simple features. The largest component of the package is the data for the two built-in atlases. Plotting results of analyses on regions or networks often involves swapping between statistical tools, like R, and software for brain imaging to correctly visualise analysis results. This package aims to make it possible to plot results directly through R.”
icon-github-alt Source URL: https://github.com/ggseg/ggseg.git
citHeader("To cite ggseg in publications use:")
citEntry(
entry = "misc",
title = "Visualisation of Brain Statistics with R-packages ggseg and ggseg3d",
author = "Athanasia M. Mowinckel and Didac Vidal-Piñeiro",
year = "2019",
eprint = "1912.08200",
archivePrefix = "arXiv",
primaryClass = "stat.OT",
textVersion = paste("Athanasia M. Mowinckel and Didac Vidal-Piñeiro (2019).",
"Visualisation of Brain Statistics with R-packages ggseg and ggseg3d.",
"arXiv:1912.08200")
)
 The ggseg project is funded by European Union Horizon 2020 Grant “Healthy minds 0-100 years: Optimizing the use of European brain imaging cohorts (Lifebrain),” with grant agreement 732592 (DOI: 10.3030/732592).
The ggseg project is funded by European Union Horizon 2020 Grant “Healthy minds 0-100 years: Optimizing the use of European brain imaging cohorts (Lifebrain),” with grant agreement 732592 (DOI: 10.3030/732592).

=>Access SciHub via R
if (!require("remotes")) install.packages("remotes")
remotes::install_github("netique/scihubr")
scihubr::download_paper("10.1037/h0028063") #enter doi
* * *
The map is not the territory.
“The best model of a cat is another or, preferably, the same cat.” (Rosenblueth & Wiener, 1945)
Rosenblueth, A., & Wiener, N. (1945). The role of models in science. Philosophy of Science, 12, 316-321.
Website created by Dr. Christopher B. Germann (2024) | Credits
Web-stats: Page generated in 1.72966430599E+12 sec.=>UTC=>Columbus=>Linux alfa3024 5.4.34-1-pve #1 SMP PVE 5.4.34-2 (Thu, 07 May 2020 10:02:02 +0200) x86_64=>Mozilla/5.0 AppleWebKit/537.36 (KHTML, like Gecko; compatible; ClaudeBot/1.0; +claudebot@anthropic.com)








