data(ToothGrowth)
## Example plot from ?ToothGrowth
coplot(len ~ dose | supp, data = ToothGrowth, panel = panel.smooth,
xlab = "ToothGrowth data: length vs dose, given type of supplement")
## Treat dose as a factor
ToothGrowth$dose = factor(ToothGrowth$dose)
levels(ToothGrowth$dose) = c("Low", "Medium", "High")
summary(aov(len ~ supp*dose, data=ToothGrowth))
#install.packages("xtable")
library(xtable)
xtable(x, caption = NULL, label = NULL, align = NULL, digits = NULL,
display = NULL, auto = FALSE, ...)
print(xtable(d), type="html")
print(xtable(d), type="latex") # anova table to latex
#https://cran.r-project.org/web/packages/xtable/index.html
#https://rmarkdown.rstudio.com/
data(ToothGrowth) # model log2 of dose instead of dose directly ToothGrowth$dose = log2(ToothGrowth$dose) # Classical analysis for comparison lmToothGrowth <- lm(len ~ supp + dose + supp:dose, data=ToothGrowth) summary(lmToothGrowth)
x<- (1:5) y<- (1:111) pdf(file=file.choose()) hist(x) plot(x, type='o') dev.off()
Notes
file.choose() is a very handy command which saves the work associated with defining absolute and relative paths which can be quite cumbersome.
list.files for non-interactive selection.
choose.files for selecting multiple files interactively.
More
https://www.rdocumentation.org/packages/base/versions/3.5.2/topics/file.choose
# http://wiki.math.yorku.ca/index.php/VR4:_Chapter_1_summary
x <- seq( 1, 20, 0.5) # sequence from 1 to 20 by 0.5
x # print it
w <- 1 + x/2 # a weight vector
y <- x + w*rnorm(x) # generate y with standard deviations
# equal to w
dum <- data.frame( x, y, w ) # make a data frame of 3 columns
dum # typing it's name shows the object
rm( x, y, w ) # remove the original variables
fm <- lm ( y ~ x, data = dum) # fit a simple linear regression (between x and y)
summary( fm ) # and look at the analysis
fm1 <- lm( y ~ x, data = dum, weight = 1/w^2 ) # a weighted regression
summary(fm1)
lrf <- loess( y ~ x, dum) # a smooth regression curve using a
# modern local regression function
attach( dum ) # make the columns in the data frame visible
# as variables (Note: before this command, typing x and
# y would yield errors)
plot( x, y) # a standard scatterplot to which we will
# fit regression curves
lines( spline( x, fitted (lrf)), col = 2) # add in the local regression using spline
# interpolation between calculated
# points
abline(0, 1, lty = 3, col = 3) # add in the true regression line (lty=3: read line type=dotted;
# col=3: read colour=green; type "?par" for details)
abline(fm, col = 4) # add in the unweighted regression line
abline(fm1, lty = 4, col = 5) # add in the weighted regression line
plot( fitted(fm), resid(fm),
xlab = "Fitted Values",
ylab = "Residuals") # a standard regression diagnostic plot
# to check for heteroscedasticity.
# The data were generated from a
# heteroscedastic process. Can you see
# it from this plot?
qqnorm( resid(fm) ) # Normal scores to check for skewness,
# kurtosis and outliers. How would you
# expect heteroscedasticity to show up?
search() # Have a look at the search path
detach() # Remove the data frame from the search path
search() # Have another look. How did it change?
rm(fm, fm1, lrf, dum) # Clean up (this is not that important in R
# for reasons we will understand later)
library(rgl)
rgl.open()
rgl.bg(col='white')
x <- sort(rnorm(1000))
y <- rnorm(1000)
z1 <- atan2(x,y)
z <- rnorm(1000) + atan2(x, y)
plot3d(x, y, z1, col = rainbow(1000), size = 2)
bbox3d()
skew(x, na.rm = TRUE)
kurtosi(x, na.rm = TRUE)
#x A data.frame or matrix
#na.rm how to treat missing data
## The function is currently defined as
function (x, na.rm = TRUE)
{
if (length(dim(x)) == 0) {
if (na.rm) {
x <- x[!is.na(x)]
}
mx <- mean(x)
sdx <- sd(x,na.rm=na.rm)
kurt <- sum((x - mx)^4)/(length(x) * sd(x)^4) -3
} else {
kurt <- rep(NA,dim(x)[2])
mx <- colMeans(x,na.rm=na.rm)
sdx <- sd(x,na.rm=na.rm)
for (i in 1:dim(x)[2]) {
kurt[i] <- sum((x[,i] - mx[i])^4, na.rm = na.rm)/((length(x[,i]) - sum(is.na(x[,i]))) * sdx[i]^4) -3
}
}
return(kurt)
}
#https://richarddmorey.github.io/BayesFactor/ bf = ttestBF(x = diffScores) ## Equivalently: ## bf = ttestBF(x = sleep$extra[1:10],y=sleep$extra[11:20], paired=TRUE) bf
Comment
ttestBF function, which performs the “JZS” t test described by Rouder, Speckman, Sun, Morey, and Iverson (2009).
#https://richarddmorey.github.io/BayesFactor/ chains2 = recompute(chains, iterations = 10000) plot(chains2[,1:2])
Comment
The posterior function returns a object of type BFmcmc, which inherits the methods of the mcmc class from the coda package.
#https://richarddmorey.github.io/BayesFactor/ ## Compute Bayes factor bf = ttestBF(formula = weight ~ feed, data = chickwts) bf ### chains = posterior(bf, iterations = 10000) plot(chains[,2])
#https://richarddmorey.github.io/BayesFactor/ ## Bem's t statistics from four selected experiments t = c(-.15, 2.39, 2.42, 2.43) N = c(100, 150, 97, 99) ### ## Do analysis again, without nullInterval restriction bf = meta.ttestBF(t=t, n1=N, rscale=1) ## Obtain posterior samples chains = posterior(bf, iterations = 10000) plot(chains)
Comment
ttestBF function, which performs the “JZS” t test described by Rouder, Speckman, Sun, Morey, and Iverson (2009).
## This source code is licensed under the FreeBSD license
## (c) 2013 Felix Schönbrodt
#' @title Plots a comparison of a sequence of priors for t test Bayes factors
#'
#' @details
#'
#'
#' @param ts A vector of t values
#' @param ns A vector of corresponding sample sizes
#' @param rs The sequence of rs that should be tested. r should run up to 2 (higher values are implausible; E.-J. Wagenmakers, personal communication, Aug 22, 2013)
#' @param labels Names for the studies (displayed in the facet headings)
#' @param dots Values of r's which should be marked with a red dot
#' @param plot If TRUE, a ggplot is returned. If false, a data frame with the computed Bayes factors is returned
#' @param sides If set to "two" (default), a two-sided Bayes factor is computed. If set to "one", a one-sided Bayes factor is computed. In this case, it is assumed that positive t values correspond to results in the predicted direction and negative t values to results in the unpredicted direction. For details, see Wagenmakers, E. J., & Morey, R. D. (2013). Simple relation between one-sided and two-sided Bayesian point-null hypothesis tests.
#' @param nrow Number of rows of the faceted plot.
#' @param forH1 Defines the direction of the BF. If forH1 is TRUE, BF > 1 speak in favor of H1 (i.e., the quotient is defined as H1/H0). If forH1 is FALSE, it's the reverse direction.
#'
#' @references
#'
#' Rouder, J. N., Speckman, P. L., Sun, D., Morey, R. D., & Iverson, G. (2009). Bayesian t-tests for accepting and rejecting the null hypothesis. Psychonomic Bulletin and Review, 16, 225-237.
#' Wagenmakers, E.-J., & Morey, R. D. (2013). Simple relation between one-sided and two-sided Bayesian point-null hypothesis tests. Manuscript submitted for publication
#' Wagenmakers, E.-J., Wetzels, R., Borsboom, D., Kievit, R. & van der Maas, H. L. J. (2011). Yes, psychologists must change the way they analyze their data: Clarifications for Bem, Utts, & Johnson (2011)
BFrobustplot <- function(
ts, ns, rs=seq(0, 2, length.out=200), dots=1, plot=TRUE,
labels=c(), sides="two", nrow=2, xticks=3, forH1=TRUE)
{
library(BayesFactor)
# compute one-sided p-values from ts and ns
ps <- pt(ts, df=ns-1, lower.tail = FALSE) # one-sided test
# add the dots location to the sequences of r's
rs <- c(rs, dots)
res <- data.frame()
for (r in rs) {
# first: calculate two-sided BF
B_e0 <- c()
for (i in 1:length(ts))
B_e0 <- c(B_e0, exp(ttest.tstat(t = ts[i], n1 = ns[i], rscale=r)$bf))
# second: calculate one-sided BF
B_r0 <- c() for (i in 1:length(ts)) { if (ts[i] > 0) {
# correct direction
B_r0 <- c(B_r0, (2 - 2*ps[i])*B_e0[i])
} else {
# wrong direction
B_r0 <- c(B_r0, (1 - ps[i])*2*B_e0[i])
}
}
res0 <- data.frame(t=ts, n=ns, BF_two=B_e0, BF_one=B_r0, r=r) if (length(labels) > 0) {
res0$labels <- labels
res0$heading <- factor(1:length(labels), labels=paste0(labels, "\n(t = ", ts, ", df = ", ns-1, ")"), ordered=TRUE)
} else {
res0$heading <- factor(1:length(ts), labels=paste0("t = ", ts, ", df = ", ns-1), ordered=TRUE)
}
res <- rbind(res, res0)
}
# define the measure to be plotted: one- or two-sided?
res$BF <- res[, paste0("BF_", sides)]
# Flip BF if requested
if (forH1 == FALSE) {
res$BF <- 1/res$BF
}
if (plot==TRUE) {
library(ggplot2)
p1 <- ggplot(res, aes(x=r, y=log(BF))) + geom_line() + facet_wrap(~heading, nrow=nrow) + theme_bw() + ylab("log(BF)")
p1 <- p1 + geom_hline(yintercept=c(c(-log(c(30, 10, 3)), log(c(3, 10, 30)))), linetype="dotted", color="darkgrey")
p1 <- p1 + geom_hline(yintercept=log(1), linetype="dashed", color="darkgreen")
# add the dots
p1 <- p1 + geom_point(data=res[res$r %in% dots,], aes(x=r, y=log(BF)), color="red", size=2)
# add annotation
p1 <- p1 + annotate("text", x=max(rs)*1.8, y=-2.85, label=paste0("Strong~H[", ifelse(forH1==TRUE,0,1), "]"), hjust=1, vjust=.5, size=3, color="black", parse=TRUE)
p1 <- p1 + annotate("text", x=max(rs)*1.8, y=-1.7 , label=paste0("Moderate~H[", ifelse(forH1==TRUE,0,1), "]"), hjust=1, vjust=.5, size=3, color="black", parse=TRUE)
p1 <- p1 + annotate("text", x=max(rs)*1.8, y=-.55 , label=paste0("Anectodal~H[", ifelse(forH1==TRUE,0,1), "]"), hjust=1, vjust=.5, size=3, color="black", parse=TRUE)
p1 <- p1 + annotate("text", x=max(rs)*1.8, y=2.86 , label=paste0("Strong~H[", ifelse(forH1==TRUE,1,0), "]"), hjust=1, vjust=.5, size=3, color="black", parse=TRUE)
p1 <- p1 + annotate("text", x=max(rs)*1.8, y=1.7 , label=paste0("Moderate~H[", ifelse(forH1==TRUE,1,0), "]"), hjust=1, vjust=.5, size=3, color="black", parse=TRUE)
p1 <- p1 + annotate("text", x=max(rs)*1.8, y=.55 , label=paste0("Anectodal~H[", ifelse(forH1==TRUE,1,0), "]"), hjust=1, vjust=.5, vjust=.5, size=3, color="black", parse=TRUE)
# set scale ticks
p1 <- p1 + scale_y_continuous(breaks=c(c(-log(c(30, 10, 3)), 0, log(c(3, 10, 30)))), labels=c("-log(30)", "-log(10)", "-log(3)", "log(1)", "log(3)", "log(10)", "log(30)"))
p1 <- p1 + scale_x_continuous(breaks=seq(min(rs), max(rs), length.out=xticks))
return(p1)
} else {
return(res)
}
}
References
Rouder, J. N., Speckman, P. L., Sun, D., Morey, R. D., & Iverson, G. (2009). Bayesian t-tests for accepting and rejecting the null hypothesis. Psychonomic Bulletin and Review, 16, 225-237
#https://jimgrange.wordpress.com/2015/11/27/animating-robustness-check-of-bayes-factor-priors/
### plots of prior robustness check
# gif created from PNG plots using http://gifmaker.me/
# clear R's memory
rm(list = ls())
# load the Bayes factor package
library(BayesFactor)
# set working directory (will be used to save files here, so make sure
# this is where you want to save your plots!)
setwd <- "D:/Work//Blog_YouTube code//Blog/Prior Robust Visualisation/plots"
#------------------------------------------------------------------------------
### declare some variables for the analysis
# what is the t-value for the data?
tVal <- 3.098
# how many points in the prior should be explored?
nPoints <- 1000
# what Cauchy rates should be explored?
cauchyRates <- seq(from = 0.01, to = 1.5, length.out = nPoints)
# what effect sizes should be plotted?
effSize <- seq(from = -2, to = 2, length.out = nPoints)
# get the Bayes factor for each prior value
bayesFactors <- sapply(cauchyRates, function(x)
exp(ttest.tstat(t = tVal, n1 = 76, rscale = x)[['bf']]))
#------------------------------------------------------------------------------
#------------------------------------------------------------------------------
### do the plotting
# how many plots do we want to produce?
nPlots <- 50
plotWidth <- round(seq(from = 1, to = nPoints, length.out = nPlots), 0)
# loop over each plot
for(i in plotWidth){
# set up the file
currFile <- paste(getwd(), "/plot_", i, ".png", sep = "")
# initiate the png file
png(file = currFile, width = 1200, height = 1000, res = 200)
# change the plotting window so plots appear side-by-side
par(mfrow = c(1, 2))
#----
# do the prior density plot
d <- dcauchy(effSize, scale = cauchyRates[i])
plot(effSize, d, type = "l", ylim = c(0, 5), xlim = c(-2, 2),
ylab = "Density", xlab = "Effect Size (d)", lwd = 2, col = "gray48",
main = paste("Rate (r) = ", round(cauchyRates[i], 3), sep = ""))
#-----
# do the Bayes factor plot
plot(cauchyRates, bayesFactors, type = "l", lwd = 2, col = "gray48",
ylim = c(0, max(bayesFactors)), xaxt = "n",
xlab = "Cauchy Prior Width (r)", ylab = "Bayes Factor (10)")
abline(h = 0, lwd = 1)
abline(h = 6, col = "black", lty = 2, lwd = 2)
axis(1, at = seq(0, 1.5, 0.25))
# add the BF at the default Cauchy point
points(0.707, 9.97, col = "black", cex = 1.5, pch = 21, bg = "red")
# add the BF for the Cauchy prior currently being plotted
points(cauchyRates[i], bayesFactors[i], col = "black", pch = 21, cex = 1.3,
bg = "cyan")
# add legend
legend(x = 0.25, y = 3, legend = c("r = 0.707", paste("r = ",
round(cauchyRates[i], 3),
sep = "")),
pch = c(21, 21), lty = c(NA, NA), lwd = c(NA, NA), pt.cex = c(1, 1),
col = c("black", "black"), pt.bg = c("red", "cyan"), bty = "n")
# save the current plot
dev.off()
}
#------------------------------------------------------------------------------
skew(x, na.rm = TRUE)
kurtosi(x, na.rm = TRUE)
#x A data.frame or matrix
#na.rm how to treat missing data
## The function is currently defined as
function (x, na.rm = TRUE)
{
if (length(dim(x)) == 0) {
if (na.rm) {
x <- x[!is.na(x)]
}
mx <- mean(x)
sdx <- sd(x,na.rm=na.rm)
kurt <- sum((x - mx)^4)/(length(x) * sd(x)^4) -3
} else {
kurt <- rep(NA,dim(x)[2])
mx <- colMeans(x,na.rm=na.rm)
sdx <- sd(x,na.rm=na.rm)
for (i in 1:dim(x)[2]) {
kurt[i] <- sum((x[,i] - mx[i])^4, na.rm = na.rm)/((length(x[,i]) - sum(is.na(x[,i]))) * sdx[i]^4) -3
}
}
return(kurt)
}
#https://richarddmorey.github.io/BayesFactor/
data(ToothGrowth)
## Example plot from ?ToothGrowth
coplot(len ~ dose | supp, data = ToothGrowth, panel = panel.smooth,
xlab = "ToothGrowth data: length vs dose, given type of supplement")
bf = anovaBF(len ~ supp*dose, data=ToothGrowth)
bf
plot(bf[3:4] / bf[2])
#reduce number of comparisons (alpha inflation)
bf = anovaBF(len ~ supp*dose, data=ToothGrowth, whichModels="top")
bf
bfMainEffects = lmBF(len ~ supp + dose, data = ToothGrowth) bfInteraction = lmBF(len ~ supp + dose + supp:dose, data = ToothGrowth) ## Compare the two models bf = bfInteraction / bfMainEffects bf ## newbf = recompute(bf, iterations = 500000) newbf # posterior function ## Sample from the posterior of the full model chains = posterior(bfInteraction, iterations = 10000) ## 1:13 are the only "interesting" parameters summary(chains[,1:13]) #lot posterior of selected effects plot(chains[,4:6])
Comment
Reduce number of comparisons by specifyig the xact model of interest a priori.
# MODIFIED FROM BEST.R FOR ONE GROUP INSTEAD OF TWO.
# Version of Dec 02, 2015.
# John K. Kruschke
# johnkruschke@gmail.com
# http://www.indiana.edu/~kruschke/BEST/
#
# This program is believed to be free of errors, but it comes with no guarantee!
# The user bears all responsibility for interpreting the results.
# Please check the webpage above for updates or corrections.
#
### ***************************************************************
### ******** SEE FILE BEST1Gexample.R FOR INSTRUCTIONS ************
### ***************************************************************
source("openGraphSaveGraph.R") # graphics functions for Windows, Mac OS, Linux
BEST1Gmcmc = function( y , numSavedSteps=100000, thinSteps=1, showMCMC=FALSE) {
# This function generates an MCMC sample from the posterior distribution.
# Description of arguments:
# showMCMC is a flag for displaying diagnostic graphs of the chains.
# If F (the default), no chain graphs are displayed. If T, they are.
require(rjags)
#------------------------------------------------------------------------------
# THE MODEL.
modelString = "
model {
for ( i in 1:Ntotal ) {
y[i] ~ dt( mu , tau , nu )
}
mu ~ dnorm( muM , muP )
tau <- 1/pow( sigma , 2 )
sigma ~ dunif( sigmaLow , sigmaHigh )
nu ~ dexp(1/30)
}
" # close quote for modelString
# Write out modelString to a text file
writeLines( modelString , con="BESTmodel.txt" )
#------------------------------------------------------------------------------
# THE DATA.
# Load the data:
Ntotal = length(y)
# Specify the data in a list, for later shipment to JAGS:
dataList = list(
y = y ,
Ntotal = Ntotal ,
muM = mean(y) ,
muP = 0.000001 * 1/sd(y)^2 ,
sigmaLow = sd(y) / 1000 ,
sigmaHigh = sd(y) * 1000
)
#------------------------------------------------------------------------------
# INTIALIZE THE CHAINS.
# Initial values of MCMC chains based on data:
mu = mean(y)
sigma = sd(y)
# Regarding initial values in next line: (1) sigma will tend to be too big if
# the data have outliers, and (2) nu starts at 5 as a moderate value. These
# initial values keep the burn-in period moderate.
initsList = list( mu = mu , sigma = sigma , nu = 5 )
#------------------------------------------------------------------------------
# RUN THE CHAINS
parameters = c( "mu" , "sigma" , "nu" ) # The parameters to be monitored
adaptSteps = 500 # Number of steps to "tune" the samplers
burnInSteps = 1000
nChains = 3
nIter = ceiling( ( numSavedSteps * thinSteps ) / nChains )
# Create, initialize, and adapt the model:
jagsModel = jags.model( "BESTmodel.txt" , data=dataList , inits=initsList ,
n.chains=nChains , n.adapt=adaptSteps )
# Burn-in:
cat( "Burning in the MCMC chain...\n" )
update( jagsModel , n.iter=burnInSteps )
# The saved MCMC chain:
cat( "Sampling final MCMC chain...\n" )
codaSamples = coda.samples( jagsModel , variable.names=parameters ,
n.iter=nIter , thin=thinSteps )
# resulting codaSamples object has these indices:
# codaSamples[[ chainIdx ]][ stepIdx , paramIdx ]
#------------------------------------------------------------------------------
# EXAMINE THE RESULTS
if ( showMCMC ) {
openGraph(width=7,height=7)
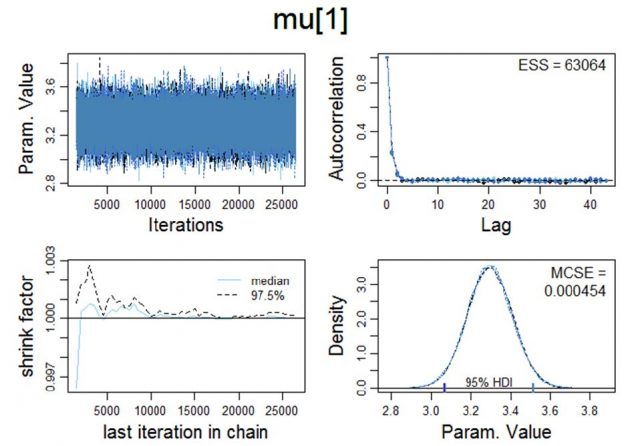
autocorr.plot( codaSamples[[1]] , ask=FALSE )
show( gelman.diag( codaSamples ) )
effectiveChainLength = effectiveSize( codaSamples )
show( effectiveChainLength )
}
# Convert coda-object codaSamples to matrix object for easier handling.
# But note that this concatenates the different chains into one long chain.
# Result is mcmcChain[ stepIdx , paramIdx ]
mcmcChain = as.matrix( codaSamples )
return( mcmcChain )
} # end function BESTmcmc
#==============================================================================
BEST1Gplot = function( y , mcmcChain , compValm=0.0 , ROPEm=NULL ,
compValsd=NULL , ROPEsd=NULL ,
ROPEeff=NULL , showCurve=FALSE , pairsPlot=FALSE ) {
# This function plots the posterior distribution (and data).
# Description of arguments:
# y is the data vector.
# mcmcChain is a list of the type returned by function BEST1G.
# compValm is hypothesized mean for comparing with estimated mean.
# ROPEm is a two element vector, such as c(-1,1), specifying the limit
# of the ROPE on the mean.
# ROPEsd is a two element vector, such as c(14,16), specifying the limit
# of the ROPE on the difference of standard deviations.
# ROPEeff is a two element vector, such as c(-1,1), specifying the limit
# of the ROPE on the effect size.
# showCurve is TRUE or FALSE and indicates whether the posterior should
# be displayed as a histogram (by default) or by an approximate curve.
# pairsPlot is TRUE or FALSE and indicates whether scatterplots of pairs
# of parameters should be displayed.
mu = mcmcChain[,"mu"]
sigma = mcmcChain[,"sigma"]
nu = mcmcChain[,"nu"]
if ( pairsPlot ) {
# Plot the parameters pairwise, to see correlations:
openGraph(width=7,height=7)
nPtToPlot = 1000
plotIdx = floor(seq(1,length(mu),by=length(mu)/nPtToPlot))
panel.cor = function(x, y, digits=2, prefix="", cex.cor, ...) {
usr = par("usr"); on.exit(par(usr))
par(usr = c(0, 1, 0, 1))
r = (cor(x, y))
txt = format(c(r, 0.123456789), digits=digits)[1]
txt = paste(prefix, txt, sep="")
if(missing(cex.cor)) cex.cor <- 0.8/strwidth(txt)
text(0.5, 0.5, txt, cex=1.25 ) # was cex=cex.cor*r
}
pairs( cbind( mu , sigma , log10(nu) )[plotIdx,] ,
labels=c( expression(mu) ,
expression(sigma) ,
expression(log10(nu)) ) ,
lower.panel=panel.cor , col="skyblue" )
}
# Define matrix for storing summary info:
summaryInfo = NULL
source("plotPost.R")
# Set up window and layout:
openGraph(width=6.0,height=5.0)
layout( matrix( c(3,3,4,4,5,5, 1,1,1,1,2,2) , nrow=6, ncol=2 , byrow=FALSE ) )
par( mar=c(3.5,3.5,2.5,0.5) , mgp=c(2.25,0.7,0) )
# Select thinned steps in chain for plotting of posterior predictive curves:
chainLength = NROW( mcmcChain )
nCurvesToPlot = 30
stepIdxVec = seq( 1 , chainLength , floor(chainLength/nCurvesToPlot) )
xRange = range( y )
xLim = c( xRange[1]-0.1*(xRange[2]-xRange[1]) ,
xRange[2]+0.1*(xRange[2]-xRange[1]) )
xVec = seq( xLim[1] , xLim[2] , length=200 )
maxY = max( dt( 0 , df=max(nu[stepIdxVec]) ) / min(sigma[stepIdxVec]) )
# Plot data y and smattering of posterior predictive curves:
stepIdx = 1
plot( xVec , dt( (xVec-mu[stepIdxVec[stepIdx]])/sigma[stepIdxVec[stepIdx]] ,
df=nu[stepIdxVec[stepIdx]] )/sigma[stepIdxVec[stepIdx]] ,
ylim=c(0,maxY) , cex.lab=1.75 ,
type="l" , col="skyblue" , lwd=1 , xlab="y" , ylab="p(y)" ,
main="Data w. Post. Pred." )
for ( stepIdx in 2:length(stepIdxVec) ) {
lines(xVec, dt( (xVec-mu[stepIdxVec[stepIdx]])/sigma[stepIdxVec[stepIdx]] ,
df=nu[stepIdxVec[stepIdx]] )/sigma[stepIdxVec[stepIdx]] ,
type="l" , col="skyblue" , lwd=1 )
}
histBinWd = median(sigma)/2
histCenter = mean(mu)
histBreaks = sort( c( seq( histCenter-histBinWd/2 , min(xVec)-histBinWd/2 ,
-histBinWd ),
seq( histCenter+histBinWd/2 , max(xVec)+histBinWd/2 ,
histBinWd ) , xLim ) )
histInfo = hist( y , plot=FALSE , breaks=histBreaks )
yPlotVec = histInfo$density
yPlotVec[ yPlotVec==0.0 ] = NA
xPlotVec = histInfo$mids
xPlotVec[ yPlotVec==0.0 ] = NA
points( xPlotVec , yPlotVec , type="h" , lwd=3 , col="red" )
text( max(xVec) , maxY , bquote(N==.(length(y))) , adj=c(1.1,1.1) )
# Plot posterior distribution of parameter nu:
paramInfo = plotPost( log10(nu) , col="skyblue" , # breaks=30 ,
showCurve=showCurve ,
xlab=bquote("log10("*nu*")") , cex.lab = 1.75 , showMode=TRUE ,
main="Normality" ) # (<0.7 suggests kurtosis)
summaryInfo = rbind( summaryInfo , paramInfo )
rownames(summaryInfo)[NROW(summaryInfo)] = "log10(nu)"
# Plot posterior distribution of parameters mu1, mu2, and their difference:
xlim = range( c( mu , compValm ) )
paramInfo = plotPost( mu , xlim=xlim , cex.lab = 1.75 , ROPE=ROPEm ,
showCurve=showCurve , compVal = compValm ,
xlab=bquote(mu) , main=paste("Mean") ,
col="skyblue" )
summaryInfo = rbind( summaryInfo , paramInfo )
rownames(summaryInfo)[NROW(summaryInfo)] = "mu"
# Plot posterior distribution of sigma:
xlim=range( sigma )
paramInfo = plotPost( sigma , xlim=xlim , cex.lab = 1.75 , ROPE=ROPEsd ,
showCurve=showCurve , compVal=compValsd ,
xlab=bquote(sigma) , main=paste("Std. Dev.") ,
col="skyblue" , showMode=TRUE )
summaryInfo = rbind( summaryInfo , paramInfo )
rownames(summaryInfo)[NROW(summaryInfo)] = "sigma"
# Plot of estimated effect size:
effectSize = ( mu - compValm ) / sigma
paramInfo = plotPost( effectSize , compVal=0 , ROPE=ROPEeff ,
showCurve=showCurve ,
xlab=bquote( (mu-.(compValm)) / sigma ),
showMode=TRUE , cex.lab=1.75 , main="Effect Size" , col="skyblue" )
summaryInfo = rbind( summaryInfo , paramInfo )
rownames(summaryInfo)[NROW(summaryInfo)] = "effSz"
return( summaryInfo )
} # end of function BEST1Gplot
#==============================================================================
# MODIFIED 2015 DEC 02.
# John K. Kruschke
# johnkruschke@gmail.com
# http://www.indiana.edu/~kruschke/BEST/
#
# This program is believed to be free of errors, but it comes with no guarantee!
# The user bears all responsibility for interpreting the results.
# Please check the webpage above for updates or corrections.
#
### ***************************************************************
### ******** SEE FILE BESTexample.R FOR INSTRUCTIONS **************
### ***************************************************************
# Load various essential functions:
source("DBDA2E-utilities.R")
BESTmcmc = function( y1 , y2 ,
priorOnly=FALSE , showMCMC=FALSE ,
numSavedSteps=20000 , thinSteps=1 ,
mu1PriorMean = mean(c(y1,y2)) ,
mu1PriorSD = sd(c(y1,y2))*5 ,
mu2PriorMean = mean(c(y1,y2)) ,
mu2PriorSD = sd(c(y1,y2))*5 ,
sigma1PriorMode = sd(c(y1,y2)) ,
sigma1PriorSD = sd(c(y1,y2))*5 ,
sigma2PriorMode = sd(c(y1,y2)) ,
sigma2PriorSD = sd(c(y1,y2))*5 ,
nuPriorMean = 30 ,
nuPriorSD = 30 ,
runjagsMethod=runjagsMethodDefault ,
nChains=nChainsDefault ) {
# This function generates an MCMC sample from the posterior distribution.
# Description of arguments:
# showMCMC is a flag for displaying diagnostic graphs of the chains.
# If F (the default), no chain graphs are displayed. If T, they are.
#------------------------------------------------------------------------------
# THE DATA.
# Load the data:
y = c( y1 , y2 ) # combine data into one vector
x = c( rep(1,length(y1)) , rep(2,length(y2)) ) # create group membership code
Ntotal = length(y)
# Specify the data and prior constants in a list, for later shipment to JAGS:
if ( priorOnly ) {
dataList = list(
# y = y ,
x = x ,
Ntotal = Ntotal ,
mu1PriorMean = mu1PriorMean ,
mu1PriorSD = mu1PriorSD ,
mu2PriorMean = mu2PriorMean ,
mu2PriorSD = mu2PriorSD ,
Sh1 = gammaShRaFromModeSD( mode=sigma1PriorMode ,
sd=sigma1PriorSD )$shape ,
Ra1 = gammaShRaFromModeSD( mode=sigma1PriorMode ,
sd=sigma1PriorSD )$rate ,
Sh2 = gammaShRaFromModeSD( mode=sigma2PriorMode ,
sd=sigma2PriorSD )$shape ,
Ra2 = gammaShRaFromModeSD( mode=sigma2PriorMode ,
sd=sigma2PriorSD )$rate ,
ShNu = gammaShRaFromMeanSD( mean=nuPriorMean , sd=nuPriorSD )$shape ,
RaNu = gammaShRaFromMeanSD( mean=nuPriorMean , sd=nuPriorSD )$rate
)
} else {
dataList = list(
y = y ,
x = x ,
Ntotal = Ntotal ,
mu1PriorMean = mu1PriorMean ,
mu1PriorSD = mu1PriorSD ,
mu2PriorMean = mu2PriorMean ,
mu2PriorSD = mu2PriorSD ,
Sh1 = gammaShRaFromModeSD( mode=sigma1PriorMode ,
sd=sigma1PriorSD )$shape ,
Ra1 = gammaShRaFromModeSD( mode=sigma1PriorMode ,
sd=sigma1PriorSD )$rate ,
Sh2 = gammaShRaFromModeSD( mode=sigma2PriorMode ,
sd=sigma2PriorSD )$shape ,
Ra2 = gammaShRaFromModeSD( mode=sigma2PriorMode ,
sd=sigma2PriorSD )$rate ,
ShNu = gammaShRaFromMeanSD( mean=nuPriorMean , sd=nuPriorSD )$shape ,
RaNu = gammaShRaFromMeanSD( mean=nuPriorMean , sd=nuPriorSD )$rate
)
}
#----------------------------------------------------------------------------
# THE MODEL.
modelString = "
model {
for ( i in 1:Ntotal ) {
y[i] ~ dt( mu[x[i]] , 1/sigma[x[i]]^2 , nu )
}
mu[1] ~ dnorm( mu1PriorMean , 1/mu1PriorSD^2 ) # prior for mu[1]
sigma[1] ~ dgamma( Sh1 , Ra1 ) # prior for sigma[1]
mu[2] ~ dnorm( mu2PriorMean , 1/mu2PriorSD^2 ) # prior for mu[2]
sigma[2] ~ dgamma( Sh2 , Ra2 ) # prior for sigma[2]
nu ~ dgamma( ShNu , RaNu ) # prior for nu
}
" # close quote for modelString
# Write out modelString to a text file
writeLines( modelString , con="BESTmodel.txt" )
#------------------------------------------------------------------------------
# INTIALIZE THE CHAINS.
# Initial values of MCMC chains based on data:
mu = c( mean(y1) , mean(y2) )
sigma = c( sd(y1) , sd(y2) )
# Regarding initial values in next line: (1) sigma will tend to be too big if
# the data have outliers, and (2) nu starts at 5 as a moderate value. These
# initial values keep the burn-in period moderate.
initsList = list( mu = mu , sigma = sigma , nu = 5 )
#------------------------------------------------------------------------------
# RUN THE CHAINS
parameters = c( "mu" , "sigma" , "nu" ) # The parameters to be monitored
adaptSteps = 500 # Number of steps to "tune" the samplers
burnInSteps = 1000
runJagsOut <- run.jags( method=runjagsMethod , model="BESTmodel.txt" , monitor=parameters , data=dataList , inits=initsList , n.chains=nChains , adapt=adaptSteps , burnin=burnInSteps , sample=ceiling(numSavedSteps/nChains) , thin=thinSteps , summarise=FALSE , plots=FALSE ) codaSamples = as.mcmc.list( runJagsOut ) # resulting codaSamples object has these indices: # codaSamples[[ chainIdx ]][ stepIdx , paramIdx ] ## Original version, using rjags: # nChains = 3 # nIter = ceiling( ( numSavedSteps * thinSteps ) / nChains ) # # Create, initialize, and adapt the model: # jagsModel = jags.model( "BESTmodel.txt" , data=dataList , inits=initsList , # n.chains=nChains , n.adapt=adaptSteps ) # # Burn-in: # cat( "Burning in the MCMC chain...\n" ) # update( jagsModel , n.iter=burnInSteps ) # # The saved MCMC chain: # cat( "Sampling final MCMC chain...\n" ) # codaSamples = coda.samples( jagsModel , variable.names=parameters , # n.iter=nIter , thin=thinSteps ) #------------------------------------------------------------------------------ # EXAMINE THE RESULTS if ( showMCMC ) { for ( paramName in varnames(codaSamples) ) { diagMCMC( codaSamples , parName=paramName ) } } # Convert coda-object codaSamples to matrix object for easier handling. # But note that this concatenates the different chains into one long chain. # Result is mcmcChain[ stepIdx , paramIdx ] mcmcChain = as.matrix( codaSamples ) return( mcmcChain ) } # end function BESTmcmc #============================================================================== BESTsummary = function( y1 , y2 , mcmcChain ) { #source("HDIofMCMC.R") # in DBDA2E-utilities.R mcmcSummary = function( paramSampleVec , compVal=NULL ) { meanParam = mean( paramSampleVec ) medianParam = median( paramSampleVec ) dres = density( paramSampleVec ) modeParam = dres$x[which.max(dres$y)] hdiLim = HDIofMCMC( paramSampleVec ) if ( !is.null(compVal) ) { pcgtCompVal = ( 100 * sum( paramSampleVec > compVal )
/ length( paramSampleVec ) )
} else {
pcgtCompVal=NA
}
return( c( meanParam , medianParam , modeParam , hdiLim , pcgtCompVal ) )
}
# Define matrix for storing summary info:
summaryInfo = matrix( 0 , nrow=9 , ncol=6 , dimnames=list(
PARAMETER=c( "mu1" , "mu2" , "muDiff" , "sigma1" , "sigma2" , "sigmaDiff" ,
"nu" , "nuLog10" , "effSz" ),
SUMMARY.INFO=c( "mean" , "median" , "mode" , "HDIlow" , "HDIhigh" ,
"pcgtZero" )
) )
summaryInfo[ "mu1" , ] = mcmcSummary( mcmcChain[,"mu[1]"] )
summaryInfo[ "mu2" , ] = mcmcSummary( mcmcChain[,"mu[2]"] )
summaryInfo[ "muDiff" , ] = mcmcSummary( mcmcChain[,"mu[1]"]
- mcmcChain[,"mu[2]"] ,
compVal=0 )
summaryInfo[ "sigma1" , ] = mcmcSummary( mcmcChain[,"sigma[1]"] )
summaryInfo[ "sigma2" , ] = mcmcSummary( mcmcChain[,"sigma[2]"] )
summaryInfo[ "sigmaDiff" , ] = mcmcSummary( mcmcChain[,"sigma[1]"]
- mcmcChain[,"sigma[2]"] ,
compVal=0 )
summaryInfo[ "nu" , ] = mcmcSummary( mcmcChain[,"nu"] )
summaryInfo[ "nuLog10" , ] = mcmcSummary( log10(mcmcChain[,"nu"]) )
N1 = length(y1)
N2 = length(y2)
effSzChain = ( ( mcmcChain[,"mu[1]"] - mcmcChain[,"mu[2]"] )
/ sqrt( ( mcmcChain[,"sigma[1]"]^2
+ mcmcChain[,"sigma[2]"]^2 ) / 2 ) )
summaryInfo[ "effSz" , ] = mcmcSummary( effSzChain , compVal=0 )
# Or, use sample-size weighted version:
# effSz = ( mu1 - mu2 ) / sqrt( ( sigma1^2 *(N1-1) + sigma2^2 *(N2-1) )
# / (N1+N2-2) )
# Be sure also to change plot label in BESTplot function, below.
return( summaryInfo )
}
#==============================================================================
BESTplot = function( y1 , y2 , mcmcChain , ROPEm=NULL , ROPEsd=NULL ,
ROPEeff=NULL , showCurve=FALSE , pairsPlot=FALSE ) {
# This function plots the posterior distribution (and data).
# Description of arguments:
# y1 and y2 are the data vectors.
# mcmcChain is a list of the type returned by function BTT.
# ROPEm is a two element vector, such as c(-1,1), specifying the limit
# of the ROPE on the difference of means.
# ROPEsd is a two element vector, such as c(-1,1), specifying the limit
# of the ROPE on the difference of standard deviations.
# ROPEeff is a two element vector, such as c(-1,1), specifying the limit
# of the ROPE on the effect size.
# showCurve is TRUE or FALSE and indicates whether the posterior should
# be displayed as a histogram (by default) or by an approximate curve.
# pairsPlot is TRUE or FALSE and indicates whether scatterplots of pairs
# of parameters should be displayed.
mu1 = mcmcChain[,"mu[1]"]
mu2 = mcmcChain[,"mu[2]"]
sigma1 = mcmcChain[,"sigma[1]"]
sigma2 = mcmcChain[,"sigma[2]"]
nu = mcmcChain[,"nu"]
if ( pairsPlot ) {
# Plot the parameters pairwise, to see correlations:
openGraph(width=7,height=7)
nPtToPlot = 1000
plotIdx = floor(seq(1,length(mu1),by=length(mu1)/nPtToPlot))
panel.cor = function(x, y, digits=2, prefix="", cex.cor, ...) {
usr = par("usr"); on.exit(par(usr))
par(usr = c(0, 1, 0, 1))
r = (cor(x, y))
txt = format(c(r, 0.123456789), digits=digits)[1]
txt = paste(prefix, txt, sep="")
if(missing(cex.cor)) cex.cor <- 0.8/strwidth(txt)
text(0.5, 0.5, txt, cex=1.25 ) # was cex=cex.cor*r
}
pairs( cbind( mu1 , mu2 , sigma1 , sigma2 , log10(nu) )[plotIdx,] ,
labels=c( expression(mu[1]) , expression(mu[2]) ,
expression(sigma[1]) , expression(sigma[2]) ,
expression(log10(nu)) ) ,
lower.panel=panel.cor , col="skyblue" )
}
# source("plotPost.R") # in DBDA2E-utilities.R
# Set up window and layout:
openGraph(width=6.0,height=8.0)
layout( matrix( c(4,5,7,8,3,1,2,6,9,10) , nrow=5, byrow=FALSE ) )
par( mar=c(3.5,3.5,2.5,0.5) , mgp=c(2.25,0.7,0) )
# Select thinned steps in chain for plotting of posterior predictive curves:
chainLength = NROW( mcmcChain )
nCurvesToPlot = 30
stepIdxVec = seq( 1 , chainLength , floor(chainLength/nCurvesToPlot) )
xRange = range( c(y1,y2) )
if ( isTRUE( all.equal( xRange[2] , xRange[1] ) ) ) {
meanSigma = mean( c(sigma1,sigma2) )
xRange = xRange + c( -meanSigma , meanSigma )
}
xLim = c( xRange[1]-0.1*(xRange[2]-xRange[1]) ,
xRange[2]+0.1*(xRange[2]-xRange[1]) )
xVec = seq( xLim[1] , xLim[2] , length=200 )
maxY = max( dt( 0 , df=max(nu[stepIdxVec]) ) /
min(c(sigma1[stepIdxVec],sigma2[stepIdxVec])) )
# Plot data y1 and smattering of posterior predictive curves:
stepIdx = 1
plot( xVec , dt( (xVec-mu1[stepIdxVec[stepIdx]])/sigma1[stepIdxVec[stepIdx]] ,
df=nu[stepIdxVec[stepIdx]] )/sigma1[stepIdxVec[stepIdx]] ,
ylim=c(0,maxY) , cex.lab=1.75 ,
type="l" , col="skyblue" , lwd=1 , xlab="y" , ylab="p(y)" ,
main="Data Group 1 w. Post. Pred." )
for ( stepIdx in 2:length(stepIdxVec) ) {
lines(xVec, dt( (xVec-mu1[stepIdxVec[stepIdx]])/sigma1[stepIdxVec[stepIdx]] ,
df=nu[stepIdxVec[stepIdx]] )/sigma1[stepIdxVec[stepIdx]] ,
type="l" , col="skyblue" , lwd=1 )
}
histBinWd = median(sigma1)/2
histCenter = mean(mu1)
histBreaks = sort( c( seq( histCenter-histBinWd/2 , min(xVec)-histBinWd/2 ,
-histBinWd ),
seq( histCenter+histBinWd/2 , max(xVec)+histBinWd/2 ,
histBinWd ) , xLim ) )
histInfo = hist( y1 , plot=FALSE , breaks=histBreaks )
yPlotVec = histInfo$density
yPlotVec[ yPlotVec==0.0 ] = NA
xPlotVec = histInfo$mids
xPlotVec[ yPlotVec==0.0 ] = NA
points( xPlotVec , yPlotVec , type="h" , lwd=3 , col="red" )
text( max(xVec) , maxY , bquote(N[1]==.(length(y1))) , adj=c(1.1,1.1) )
# Plot data y2 and smattering of posterior predictive curves:
stepIdx = 1
plot( xVec , dt( (xVec-mu2[stepIdxVec[stepIdx]])/sigma2[stepIdxVec[stepIdx]] ,
df=nu[stepIdxVec[stepIdx]] )/sigma2[stepIdxVec[stepIdx]] ,
ylim=c(0,maxY) , cex.lab=1.75 ,
type="l" , col="skyblue" , lwd=1 , xlab="y" , ylab="p(y)" ,
main="Data Group 2 w. Post. Pred." )
for ( stepIdx in 2:length(stepIdxVec) ) {
lines(xVec, dt( (xVec-mu2[stepIdxVec[stepIdx]])/sigma2[stepIdxVec[stepIdx]] ,
df=nu[stepIdxVec[stepIdx]] )/sigma2[stepIdxVec[stepIdx]] ,
type="l" , col="skyblue" , lwd=1 )
}
histBinWd = median(sigma2)/2
histCenter = mean(mu2)
histBreaks = sort( c( seq( histCenter-histBinWd/2 , min(xVec)-histBinWd/2 ,
-histBinWd ),
seq( histCenter+histBinWd/2 , max(xVec)+histBinWd/2 ,
histBinWd ) , xLim ) )
histInfo = hist( y2 , plot=FALSE , breaks=histBreaks )
yPlotVec = histInfo$density
yPlotVec[ yPlotVec==0.0 ] = NA
xPlotVec = histInfo$mids
xPlotVec[ yPlotVec==0.0 ] = NA
points( xPlotVec , yPlotVec , type="h" , lwd=3 , col="red" )
text( max(xVec) , maxY , bquote(N[2]==.(length(y2))) , adj=c(1.1,1.1) )
# Plot posterior distribution of parameter nu:
histInfo = plotPost( log10(nu) , col="skyblue" , # breaks=30 ,
showCurve=showCurve ,
xlab=bquote("log10("*nu*")") , cex.lab = 1.75 ,
cenTend=c("mode","median","mean")[1] ,
main="Normality" ) # (<0.7 suggests kurtosis)
# Plot posterior distribution of parameters mu1, mu2, and their difference:
xlim = range( c( mu1 , mu2 ) )
histInfo = plotPost( mu1 , xlim=xlim , cex.lab = 1.75 ,
showCurve=showCurve ,
xlab=bquote(mu[1]) , main=paste("Group",1,"Mean") ,
col="skyblue" )
histInfo = plotPost( mu2 , xlim=xlim , cex.lab = 1.75 ,
showCurve=showCurve ,
xlab=bquote(mu[2]) , main=paste("Group",2,"Mean") ,
col="skyblue" )
histInfo = plotPost( mu1-mu2 , compVal=0 , showCurve=showCurve ,
xlab=bquote(mu[1] - mu[2]) , cex.lab = 1.75 , ROPE=ROPEm ,
main="Difference of Means" , col="skyblue" )
# Plot posterior distribution of param's sigma1, sigma2, and their difference:
xlim=range( c( sigma1 , sigma2 ) )
histInfo = plotPost( sigma1 , xlim=xlim , cex.lab = 1.75 ,
showCurve=showCurve ,
xlab=bquote(sigma[1]) , main=paste("Group",1,"Std. Dev.") ,
col="skyblue" , cenTend=c("mode","median","mean")[1] )
histInfo = plotPost( sigma2 , xlim=xlim , cex.lab = 1.75 ,
showCurve=showCurve ,
xlab=bquote(sigma[2]) , main=paste("Group",2,"Std. Dev.") ,
col="skyblue" , cenTend=c("mode","median","mean")[1] )
histInfo = plotPost( sigma1-sigma2 ,
compVal=0 , showCurve=showCurve ,
xlab=bquote(sigma[1] - sigma[2]) , cex.lab = 1.75 ,
ROPE=ROPEsd ,
main="Difference of Std. Dev.s" , col="skyblue" ,
cenTend=c("mode","median","mean")[1] )
# Plot of estimated effect size. Effect size is d-sub-a from
# Macmillan & Creelman, 1991; Simpson & Fitter, 1973; Swets, 1986a, 1986b.
effectSize = ( mu1 - mu2 ) / sqrt( ( sigma1^2 + sigma2^2 ) / 2 )
histInfo = plotPost( effectSize , compVal=0 , ROPE=ROPEeff ,
showCurve=showCurve ,
xlab=bquote( (mu[1]-mu[2])
/sqrt((sigma[1]^2 +sigma[2]^2 )/2 ) ),
cenTend=c("mode","median","mean")[1] , cex.lab=1.0 , main="Effect Size" ,
col="skyblue" )
# Or use sample-size weighted version:
# Hedges 1981; Wetzels, Raaijmakers, Jakab & Wagenmakers 2009.
# N1 = length(y1)
# N2 = length(y2)
# effectSize = ( mu1 - mu2 ) / sqrt( ( sigma1^2 *(N1-1) + sigma2^2 *(N2-1) )
# / (N1+N2-2) )
# Be sure also to change BESTsummary function, above.
# histInfo = plotPost( effectSize , compVal=0 , ROPE=ROPEeff ,
# showCurve=showCurve ,
# xlab=bquote( (mu[1]-mu[2])
# /sqrt((sigma[1]^2 *(N[1]-1)+sigma[2]^2 *(N[2]-1))/(N[1]+N[2]-2)) ),
# cenTend=c("mode","median","mean")[1] , cex.lab=1.0 , main="Effect Size" , col="skyblue" )
return( BESTsummary( y1 , y2 , mcmcChain ) )
} # end of function BESTplot
#==============================================================================
BESTpower = function( mcmcChain , N1 , N2 , ROPEm , ROPEsd , ROPEeff ,
maxHDIWm , maxHDIWsd , maxHDIWeff , nRep=200 ,
mcmcLength=10000 , saveName="BESTpower.Rdata" ,
showFirstNrep=0 ) {
# This function estimates power.
# Description of arguments:
# mcmcChain is a matrix of the type returned by function BESTmcmc.
# N1 and N2 are sample sizes for the two groups.
# ROPEm is a two element vector, such as c(-1,1), specifying the limit
# of the ROPE on the difference of means.
# ROPEsd is a two element vector, such as c(-1,1), specifying the limit
# of the ROPE on the difference of standard deviations.
# ROPEeff is a two element vector, such as c(-1,1), specifying the limit
# of the ROPE on the effect size.
# maxHDIWm is the maximum desired width of the 95% HDI on the difference
# of means.
# maxHDIWsd is the maximum desired width of the 95% HDI on the difference
# of standard deviations.
# maxHDIWeff is the maximum desired width of the 95% HDI on the effect size.
# nRep is the number of simulated experiments used to estimate the power.
#source("HDIofMCMC.R") # in DBDA2E-utilities.R
#source("HDIofICDF.R") # in DBDA2E-utilities.R
chainLength = NROW( mcmcChain )
# Select thinned steps in chain for posterior predictions:
stepIdxVec = seq( 1 , chainLength , floor(chainLength/nRep) )
goalTally = list( HDIm.gt.ROPE = c(0) ,
HDIm.lt.ROPE = c(0) ,
HDIm.in.ROPE = c(0) ,
HDIm.wd.max = c(0) ,
HDIsd.gt.ROPE = c(0) ,
HDIsd.lt.ROPE = c(0) ,
HDIsd.in.ROPE = c(0) ,
HDIsd.wd.max = c(0) ,
HDIeff.gt.ROPE = c(0) ,
HDIeff.lt.ROPE = c(0) ,
HDIeff.in.ROPE = c(0) ,
HDIeff.wd.max = c(0) )
power = list( HDIm.gt.ROPE = c(0,0,0) ,
HDIm.lt.ROPE = c(0,0,0) ,
HDIm.in.ROPE = c(0,0,0) ,
HDIm.wd.max = c(0,0,0) ,
HDIsd.gt.ROPE = c(0,0,0) ,
HDIsd.lt.ROPE = c(0,0,0) ,
HDIsd.in.ROPE = c(0,0,0) ,
HDIsd.wd.max = c(0,0,0) ,
HDIeff.gt.ROPE = c(0,0,0) ,
HDIeff.lt.ROPE = c(0,0,0) ,
HDIeff.in.ROPE = c(0,0,0) ,
HDIeff.wd.max = c(0,0,0) )
nSim = 0
for ( stepIdx in stepIdxVec ) {
nSim = nSim + 1
cat( "\n:::::::::::::::::::::::::::::::::::::::::::::::::::::::::::::::\n" )
cat( paste( "Power computation: Simulated Experiment" , nSim , "of" ,
length(stepIdxVec) , ":\n\n" ) )
# Get parameter values for this simulation:
mu1Val = mcmcChain[stepIdx,"mu[1]"]
mu2Val = mcmcChain[stepIdx,"mu[2]"]
sigma1Val = mcmcChain[stepIdx,"sigma[1]"]
sigma2Val = mcmcChain[stepIdx,"sigma[2]"]
nuVal = mcmcChain[stepIdx,"nu"]
# Generate simulated data:
y1 = rt( N1 , df=nuVal ) * sigma1Val + mu1Val
y2 = rt( N2 , df=nuVal ) * sigma2Val + mu2Val
# Get posterior for simulated data:
simChain = BESTmcmc( y1, y2, numSavedSteps=mcmcLength, thinSteps=1,
showMCMC=FALSE )
if (nSim <= showFirstNrep ) { BESTplot( y1, y2, simChain, ROPEm=ROPEm, ROPEsd=ROPEsd, ROPEeff=ROPEeff ) } # Assess which goals were achieved and tally them: HDIm = HDIofMCMC( simChain[,"mu[1]"] - simChain[,"mu[2]"] ) if ( HDIm[1] > ROPEm[2] ) {
goalTally$HDIm.gt.ROPE = goalTally$HDIm.gt.ROPE + 1 }
if ( HDIm[2] < ROPEm[1] ) { goalTally$HDIm.lt.ROPE = goalTally$HDIm.lt.ROPE + 1 } if ( HDIm[1] > ROPEm[1] & HDIm[2] < ROPEm[2] ) {
goalTally$HDIm.in.ROPE = goalTally$HDIm.in.ROPE + 1 }
if ( HDIm[2] - HDIm[1] < maxHDIWm ) { goalTally$HDIm.wd.max = goalTally$HDIm.wd.max + 1 } HDIsd = HDIofMCMC( simChain[,"sigma[1]"] - simChain[,"sigma[2]"] ) if ( HDIsd[1] > ROPEsd[2] ) {
goalTally$HDIsd.gt.ROPE = goalTally$HDIsd.gt.ROPE + 1 }
if ( HDIsd[2] < ROPEsd[1] ) { goalTally$HDIsd.lt.ROPE = goalTally$HDIsd.lt.ROPE + 1 } if ( HDIsd[1] > ROPEsd[1] & HDIsd[2] < ROPEsd[2] ) {
goalTally$HDIsd.in.ROPE = goalTally$HDIsd.in.ROPE + 1 }
if ( HDIsd[2] - HDIsd[1] < maxHDIWsd ) { goalTally$HDIsd.wd.max = goalTally$HDIsd.wd.max + 1 } HDIeff = HDIofMCMC( ( simChain[,"mu[1]"] - simChain[,"mu[2]"] ) / sqrt( ( simChain[,"sigma[1]"]^2 + simChain[,"sigma[2]"]^2 ) / 2 ) ) if ( HDIeff[1] > ROPEeff[2] ) {
goalTally$HDIeff.gt.ROPE = goalTally$HDIeff.gt.ROPE + 1 }
if ( HDIeff[2] < ROPEeff[1] ) { goalTally$HDIeff.lt.ROPE = goalTally$HDIeff.lt.ROPE + 1 } if ( HDIeff[1] > ROPEeff[1] & HDIeff[2] < ROPEeff[2] ) {
goalTally$HDIeff.in.ROPE = goalTally$HDIeff.in.ROPE + 1 }
if ( HDIeff[2] - HDIeff[1] < maxHDIWeff ) { goalTally$HDIeff.wd.max = goalTally$HDIeff.wd.max + 1 } for ( i in 1:length(goalTally) ) { s1 = 1 + goalTally[[i]] s2 = 1 + ( nSim - goalTally[[i]] ) power[[i]][1] = s1/(s1+s2) power[[i]][2:3] = HDIofICDF( qbeta , shape1=s1 , shape2=s2 ) } cat( "\nAfter ", nSim , " Simulated Experiments, Power Is: \n( mean, HDI_low, HDI_high )\n" ) for ( i in 1:length(power) ) { cat( names(power[i]) , round(power[[i]],3) , "\n" ) } save( nSim , power , file=saveName ) } return( power ) } # end of function BESTpower #============================================================================== makeData = function( mu1 , sd1 , mu2 , sd2 , nPerGrp , pcntOut=0 , sdOutMult=2.0 , rnd.seed=NULL ) { # Auxilliary function for generating random values from a # mixture of normal distibutions. if(!is.null(rnd.seed)){set.seed(rnd.seed)} # Set seed for random values. nOut = ceiling(nPerGrp*pcntOut/100) # Number of outliers. nIn = nPerGrp - nOut # Number from main distribution. if ( pcntOut > 100 | pcntOut < 0 ) { stop("pcntOut must be between 0 and 100.") } if ( pcntOut > 0 & pcntOut < 1 ) { warning("pcntOut is specified as percentage 0-100, not proportion 0-1.") } if ( pcntOut > 50 ) {
warning("pcntOut indicates more than 50% outliers; did you intend this?")
}
if ( nOut < 2 & pcntOut > 0 ) {
stop("Combination of nPerGrp and pcntOut yields too few outliers.")
}
if ( nIn < 2 ) { stop("Too few non-outliers.") } sdN = function( x ) { sqrt( mean( (x-mean(x))^2 ) ) } # genDat = function( mu , sigma , Nin , Nout , sigmaOutMult=sdOutMult ) { y = rnorm(n=Nin) # Random values in main distribution. y = ((y-mean(y))/sdN(y))*sigma + mu # Translate to exactly realize desired. yOut = NULL if ( Nout > 0 ) {
yOut = rnorm(n=Nout) # Random values for outliers
yOut=((yOut-mean(yOut))/sdN(yOut))*(sigma*sdOutMult)+mu # Realize exactly.
}
y = c(y,yOut) # Concatenate main with outliers.
return(y)
}
y1 = genDat( mu=mu1 , sigma=sd1 , Nin=nIn , Nout=nOut )
y2 = genDat( mu=mu2 , sigma=sd2 , Nin=nIn , Nout=nOut )
#
# Set up window and layout:
openGraph(width=7,height=7) # Plot the data.
layout(matrix(1:2,nrow=2))
histInfo = hist( y1 , main="Simulated Data" , col="pink2" , border="white" ,
xlim=range(c(y1,y2)) , breaks=30 , prob=TRUE )
text( max(c(y1,y2)) , max(histInfo$density) ,
bquote(N==.(nPerGrp)) , adj=c(1,1) )
xlow=min(histInfo$breaks)
xhi=max(histInfo$breaks)
xcomb=seq(xlow,xhi,length=1001)
lines( xcomb , dnorm(xcomb,mean=mu1,sd=sd1)*nIn/(nIn+nOut) +
dnorm(xcomb,mean=mu1,sd=sd1*sdOutMult)*nOut/(nIn+nOut) , lwd=3 )
lines( xcomb , dnorm(xcomb,mean=mu1,sd=sd1)*nIn/(nIn+nOut) ,
lty="dashed" , col="green", lwd=3)
lines( xcomb , dnorm(xcomb,mean=mu1,sd=sd1*sdOutMult)*nOut/(nIn+nOut) ,
lty="dashed" , col="red", lwd=3)
histInfo = hist( y2 , main="" , col="pink2" , border="white" ,
xlim=range(c(y1,y2)) , breaks=30 , prob=TRUE )
text( max(c(y1,y2)) , max(histInfo$density) ,
bquote(N==.(nPerGrp)) , adj=c(1,1) )
xlow=min(histInfo$breaks)
xhi=max(histInfo$breaks)
xcomb=seq(xlow,xhi,length=1001)
lines( xcomb , dnorm(xcomb,mean=mu2,sd=sd2)*nIn/(nIn+nOut) +
dnorm(xcomb,mean=mu2,sd=sd2*sdOutMult)*nOut/(nIn+nOut) , lwd=3)
lines( xcomb , dnorm(xcomb,mean=mu2,sd=sd2)*nIn/(nIn+nOut) ,
lty="dashed" , col="green", lwd=3)
lines( xcomb , dnorm(xcomb,mean=mu2,sd=sd2*sdOutMult)*nOut/(nIn+nOut) ,
lty="dashed" , col="red", lwd=3)
#
return( list( y1=y1 , y2=y2 ) )
}
#==============================================================================
# Version of May 26, 2012. Re-checked on 2015 May 08.
# John K. Kruschke
# johnkruschke@gmail.com
# http://www.indiana.edu/~kruschke/BEST/
#
# This program is believed to be free of errors, but it comes with no guarantee!
# The user bears all responsibility for interpreting the results.
# Please check the webpage above for updates or corrections.
#
### ***************************************************************
### ******** SEE FILE BESTexample.R FOR INSTRUCTIONS **************
### ***************************************************************
# OPTIONAL: Clear R's memory and graphics:
rm(list=ls()) # Careful! This clears all of R's memory!
graphics.off() # This closes all of R's graphics windows.
# Get the functions loaded into R's working memory:
source("BEST.R")
#-------------------------------------------------------------------------------
# RETROSPECTIVE POWER ANALYSIS.
# !! This section assumes you have already run BESTexample.R !!
# Re-load the saved data and MCMC chain from the previously conducted
# Bayesian analysis. This re-loads the variables y1, y2, mcmcChain, etc.
load( "BESTexampleMCMC.Rdata" )
power = BESTpower( mcmcChain , N1=length(y1) , N2=length(y2) ,
ROPEm=c(-0.1,0.1) , ROPEsd=c(-0.1,0.1) , ROPEeff=c(-0.1,0.1) ,
maxHDIWm=2.0 , maxHDIWsd=2.0 , maxHDIWeff=0.2 , nRep=1000 ,
mcmcLength=10000 , saveName = "BESTexampleRetroPower.Rdata" )
#-------------------------------------------------------------------------------
# PROSPECTIVE POWER ANALYSIS, using fictitious strong data.
# Generate large fictitious data set that expresses hypothesis:
prospectData = makeData( mu1=108, sd1=17, mu2=100, sd2=15, nPerGrp=1000,
pcntOut=10, sdOutMult=2.0, rnd.seed=NULL )
y1pro = prospectData$y1 # Merely renames simulated data for convenience below.
y2pro = prospectData$y2 # Merely renames simulated data for convenience below.
# Generate Bayesian posterior distribution from fictitious data:
# (uses fewer than usual MCMC steps because it only needs nRep credible
# parameter combinations, not a high-resolution representation)
mcmcChainPro = BESTmcmc( y1pro , y2pro , numSavedSteps=2000 )
postInfoPro = BESTplot( y1pro , y2pro , mcmcChainPro , pairsPlot=TRUE )
save( y1pro, y2pro, mcmcChainPro, postInfoPro,
file="BESTexampleProPowerMCMC.Rdata" )
# Now compute the prospective power for planned sample sizes:
N1plan = N2plan = 50 # specify planned sample size
powerPro = BESTpower( mcmcChainPro , N1=N1plan , N2=N2plan , showFirstNrep=5 ,
ROPEm=c(-1.5,1.5) , ROPEsd=c(-0.0,0.0) , ROPEeff=c(-0.0,0.0) ,
maxHDIWm=15.0 , maxHDIWsd=10.0 , maxHDIWeff=1.0 , nRep=1000 ,
mcmcLength=10000 , saveName = "BESTexampleProPower.Rdata" )
#-------------------------------------------------------------------------------
#https://cran.r-project.org/web/packages/beanplot/beanplot.pdf
par( mfrow = c( 1, 4 ) )
with(dataexp2, beanplot(v00, ylim = c(0,10), col="lightgray", main = "v00", kernel = "gaussian", cut = 3, cutmin = -Inf, cutmax = Inf, overallline = "mean", horizontal = FALSE, side = "no", jitter = NULL, beanlinewd = 2))
with(dataexp2, beanplot(v01, ylim = c(0,10), col="lightgray", main = "v01", kernel = "gaussian", cut = 3, cutmin = -Inf, cutmax = Inf, overallline = "mean", horizontal = FALSE, side = "no", jitter = NULL, beanlinewd = 2))
with(dataexp2, beanplot(v10, ylim = c(0,10), col="darkgray", main = "v10", kernel = "gaussian", cut = 3, cutmin = -Inf, cutmax = Inf, overallline = "mean", horizontal = FALSE, side = "no", jitter = NULL, beanlinewd = 2))
with(dataexp2, beanplot(v11, ylim = c(0,10), col="darkgray", main = "v11", kernel = "gaussian", cut = 3, cutmin = -Inf, cutmax = Inf, overallline = "mean", horizontal = FALSE, side = "no", jitter = NULL, beanlinewd = 2))
#with ylim and titles
with(dataexp2, beanplot(v00, ylab = "VAS Brightness rating", xlab = "beanplot showing distribution and mean",
ylim = c(0,10), col="darkgray", main = "v00", kernel = "gaussian", cut = 3, cutmin = -Inf, cutmax = Inf,
overallline = "mean", horizontal = FALSE, side = "no", jitter = NULL, beanlinewd = 2))
install.packages ("beanplot")
library("beanplot")
par( mfrow = c( 1, 2 ) )
with(dataexp2, beanplot(v00, col="darkgray", main = "v00", kernel = "gaussian", cut = 3, cutmin = -Inf, cutmax = Inf, overallline = "mean", horizontal = FALSE, side = "no", jitter = NULL, beanlinewd = 2))
with(dataexp2, beanplot(v01, col="darkgray", main = "v01", kernel = "gaussian", cut = 3, cutmin = -Inf, cutmax = Inf, overallline = "mean", horizontal = FALSE, side = "no", jitter = NULL, beanlinewd = 2))
par( mfrow = c( 1, 2 ) )
with(dataexp2, beanplot(v10, col="darkgray", main = "v10", kernel = "gaussian", cut = 3, cutmin = -Inf, cutmax = Inf, overallline = "mean", horizontal = FALSE, side = "no", jitter = NULL, beanlinewd = 2))
with(dataexp2, beanplot(v11, col="darkgray", main = "v11", kernel = "gaussian", cut = 3, cutmin = -Inf, cutmax = Inf, overallline = "mean", horizontal = FALSE, side = "no", jitter = NULL, beanlinewd = 2))
stripchart(variable ~ factor, vertical=TRUE, method="stack",
ylab="variable", data=StackedData)
#two sided
with(dataexp1, beanplot(v00, v10, col="darkgray", main = "v00 vs. v10", kernel = "gaussian", cut = 3, cutmin = -Inf, cutmax = Inf, overallline = "mean", horizontal = FALSE, side = "both", jitter = NULL, beanlinewd = 2))
#verticalpar( mfrow = c( 2, 1 ) )
install.packages ("beanplot")
library("beanplot")
par( mfrow = c( 2, 1 ) ) # layout matrix
with(dataexp1, beanplot(v00, v10, main = "v00 vs. v10", kernel = "gaussian", cut = 3, col = list("black", c("grey", "white")), xlab="VAS",
cutmin = -Inf, cutmax = Inf, overallline = "mean", horizontal = T, side = "both", jitter = NULL, beanlinewd = 2))
with(dataexp1, beanplot(v01, v11, main = "v01 vs. v11", kernel = "gaussian", cut = 3, col = list("black", c("grey", "white")), xlab="VAS",
cutmin = -Inf, cutmax = Inf, overallline = "mean", horizontal = T, side = "both", jitter = NULL, beanlinewd = 2))
#exp1
dataexp1 <-
read.table("http://www.irrational-decisions.com/phd-thesis/dataexp1.csv",
header=TRUE, sep=",", na.strings="NA", dec=".", strip.white=TRUE)
par( mfrow = c( 1, 2) )
with(dataexp1, beanplot(v00, ylim = c(0,7), col="lightgray", main = "v00", kernel = "gaussian", cut = 3, cutmin = -Inf, cutmax = Inf, overallline = "mean", horizontal = FALSE, side = "no", jitter = NULL, beanlinewd = 2))
with(dataexp1, beanplot(v10, ylim = c(0,7), col="lightgray", main = "v01", kernel = "gaussian", cut = 3, cutmin = -Inf, cutmax = Inf, overallline = "mean", horizontal = FALSE, side = "no", jitter = NULL, beanlinewd = 2))
par( mfrow = c( 1, 2) )
with(dataexp1, beanplot(v01, ylim = c(3,10), col="darkgray", main = "v10", kernel = "gaussian", cut = 3, cutmin = -Inf, cutmax = Inf, overallline = "mean", horizontal = FALSE, side = "no", jitter = NULL, beanlinewd = 2))
with(dataexp1, beanplot(v11, ylim = c(3,10), col="darkgray", main = "v11", kernel = "gaussian", cut = 3, cutmin = -Inf, cutmax = Inf, overallline = "mean", horizontal = FALSE, side = "no", jitter = NULL, beanlinewd = 2))
#histogram and rug plot plot(density(dataexp2$v00)) rug(dataexp2$v00, col=2, lwd=3.5) ############## library(MASS) plot(density(dataexp2$v00, bw=5)) rug(dataexp2$v00+ rnorm(length(dataexp2$v00), sd=5), col=2, lwd=3.5)
x = c(5) y = c(9) plot(x,y,type="h",xlim=c(0,10),ylim=c(0,10),lwd=2,col="blue",ylab="knowledge", xlab="diversity")
install.packages("gtrendsR")
library(gtrendsR)
gtrends(c("bayesian inference", "p-value"), time = "all") # since 2004
res <- gtrends(c("Bayesian inference", "Markov Chain Monte Carlo", "ANOVA"), geo = c("US", "GB", "DE", "CA",))
res <- gtrends("Markov Chain Monte Carlo", geo = c("US", "GB", "DE"))
plot(res)
#get vars
length(res)
str(res)
res$interest_over_time
ff<-res$interest_over_time
scatterplot(ff$hits~ff$date| geo, reg.line=FALSE, smooth=FALSE, spread=FALSE, boxplots=FALSE, span=0.5, ellipse=FALSE, levels=c(.5, .9), by.groups=TRUE, data=ff)
res2 <- gtrends(c("p-value", "bayes factor"))
library(plotrix)
.likelihoodShiftedT <- function(par, data) {
-sum(log(dt((data - par[1])/par[2], par[3])/par[2]))
}
.dposteriorShiftedT <- function(x, parameters, oneSided) { # function that returns the posterior density if (oneSided == FALSE) { dt((x - parameters[1])/parameters[2], parameters[3])/parameters[2] } else if (oneSided == "right") { ifelse(x >= 0, (dt((x - parameters[1])/parameters[2], parameters[3])/parameters[2])/pt((0 -
parameters[1])/parameters[2], parameters[3], lower.tail = FALSE),
0)
} else if (oneSided == "left") {
ifelse(x <= 0, (dt((x - parameters[1])/parameters[2], parameters[3])/parameters[2])/pt((0 -
parameters[1])/parameters[2], parameters[3], lower.tail = TRUE),
0)
}
}
.dprior <- function(x, r, oneSided = oneSided) {
# function that returns the prior density
if (oneSided == "right") {
y <- ifelse(x < 0, 0, 2/(pi * r * (1 + (x/r)^2)))
return(y)
}
if (oneSided == "left") {
y <- ifelse(x > 0, 0, 2/(pi * r * (1 + (x/r)^2)))
return(y)
} else {
return(1/(pi * r * (1 + (x/r)^2)))
}
}
.plotPosterior.ttest <- function(x = NULL, y = NULL, paired = FALSE, oneSided = FALSE,
BF, BFH1H0 = TRUE, iterations = 10000, rscale = "medium", lwd = 2,
cexPoints = 1.5, cexAxis = 1.2, cexYlab = 1.5, cexXlab = 1.5, cexTextBF = 1.4,
cexCI = 1.1, cexLegend = 1.2, lwdAxis = 1.2, addInformation = TRUE,
dontPlotData = FALSE) {
if (addInformation) {
par(mar = c(5.6, 5, 7, 4) + 0.1, las = 1)
} else {
par(mar = c(5.6, 5, 4, 4) + 0.1, las = 1)
}
if (dontPlotData) {
plot(1, type = "n", xlim = 0:1, ylim = 0:1, bty = "n", axes = FALSE,
xlab = "", ylab = "")
axis(1, at = 0:1, labels = FALSE, cex.axis = cexAxis, lwd = lwdAxis,
xlab = "")
axis(2, at = 0:1, labels = FALSE, cex.axis = cexAxis, lwd = lwdAxis,
ylab = "")
mtext(text = "Density", side = 2, las = 0, cex = cexYlab, line = 3.25)
mtext(expression(paste("Effect size", ~delta)), side = 1, cex = cexXlab,
line = 2.5)
return()
}
if (rscale == "medium") {
r <- sqrt(2)/2
}
if (rscale == "wide") {
r <- 1
}
if (rscale == "ultrawide") {
r <- sqrt(2)
}
if (mode(rscale) == "numeric") {
r <- rscale
}
if (oneSided == FALSE) {
nullInterval <- NULL
}
if (oneSided == "right") {
nullInterval <- c(0, Inf)
}
if (oneSided == "left") {
nullInterval <- c(-Inf, 0)
}
# sample from delta posterior
samples <- BayesFactor::ttestBF(x = x, y = y, paired = paired, posterior = TRUE,
iterations = iterations, rscale = r)
delta <- samples[, "delta"]
# fit shifted t distribution
if (is.null(y)) {
deltaHat <- mean(x)/sd(x)
N <- length(x)
df <- N - 1
sigmaStart <- 1/N
} else if (paired) {
deltaHat <- mean(x - y)/sd(x - y)
N <- length(x)
df <- N - 1
sigmaStart <- 1/N
} else if (!is.null(y) && !paired) {
N1 <- length(x)
N2 <- length(y)
sdPooled <- sqrt(((N1 - 1) * var(x) + (N2 - 1) * var(y))/(N1 +
N2))
deltaHat <- (mean(x) - mean(y))/sdPooled
df <- N1 + N2 - 2
sigmaStart <- sqrt(N1 * N2/(N1 + N2))
}
if (sigmaStart < 0.01)
sigmaStart <- 0.01
parameters <- try(silent = TRUE, expr = optim(par = c(deltaHat, sigmaStart,
df), fn = .likelihoodShiftedT, data = delta, method = "BFGS")$par)
if (class(parameters) == "try-error") {
parameters <- try(silent = TRUE, expr = optim(par = c(deltaHat,
sigmaStart, df), fn = .likelihoodShiftedT, data = delta, method = "Nelder-Mead")$par)
}
if (BFH1H0) {
BF10 <- BF
BF01 <- 1/BF10
} else {
BF01 <- BF
BF10 <- 1/BF01
}
# set limits plot
xlim <- vector("numeric", 2)
if (oneSided == FALSE) {
xlim[1] <- min(-2, quantile(delta, probs = 0.01)[[1]])
xlim[2] <- max(2, quantile(delta, probs = 0.99)[[1]])
if (length(x) < 10) {
if (addInformation) {
stretch <- 1.52
} else {
stretch <- 1.4
}
} else {
stretch <- 1.2 } } if (oneSided == "right") { # if (length(delta[delta >= 0]) < 10) return('Plotting is not
# possible: To few posterior samples in tested interval')
xlim[1] <- min(-2, quantile(delta[delta >= 0], probs = 0.01)[[1]])
xlim[2] <- max(2, quantile(delta[delta >= 0], probs = 0.99)[[1]])
if (any(is.na(xlim))) {
xlim[1] <- min(-2, .qShiftedT(0.01, parameters, oneSided = "right"))
xlim[2] <- max(2, .qShiftedT(0.99, parameters, oneSided = "right"))
}
stretch <- 1.32
}
if (oneSided == "left") {
# if (length(delta[delta <= 0]) < 10) return('Plotting is not
# possible: To few posterior samples in tested interval')
xlim[1] <- min(-2, quantile(delta[delta <= 0], probs = 0.01)[[1]])
xlim[2] <- max(2, quantile(delta[delta <= 0], probs = 0.99)[[1]])
if (any(is.na(xlim))) {
xlim[1] <- min(-2, .qShiftedT(0.01, parameters, oneSided = "left"))
xlim[2] <- max(2, .qShiftedT(0.99, parameters, oneSided = "left"))
}
stretch <- 1.32
}
xticks <- pretty(xlim)
ylim <- vector("numeric", 2)
ylim[1] <- 0
dmax <- optimize(function(x) .dposteriorShiftedT(x, parameters = parameters,
oneSided = oneSided), interval = range(xticks), maximum = TRUE)$objective
ylim[2] <- max(stretch * .dprior(0, r, oneSided = oneSided), stretch *
dmax) # get maximum density
# calculate position of 'nice' tick marks and create labels
yticks <- pretty(ylim)
xlabels <- formatC(xticks, 1, format = "f")
ylabels <- formatC(yticks, 1, format = "f")
# compute 95% credible interval & median:
if (oneSided == FALSE) {
CIlow <- quantile(delta, probs = 0.025)[[1]]
CIhigh <- quantile(delta, probs = 0.975)[[1]]
medianPosterior <- median(delta)
if (any(is.na(c(CIlow, CIhigh, medianPosterior)))) {
CIlow <- .qShiftedT(0.025, parameters, oneSided = FALSE)
CIhigh <- .qShiftedT(0.975, parameters, oneSided = FALSE)
medianPosterior <- .qShiftedT(0.5, parameters, oneSided = FALSE)
}
}
if (oneSided == "right") {
CIlow <- quantile(delta[delta >= 0], probs = 0.025)[[1]]
CIhigh <- quantile(delta[delta >= 0], probs = 0.975)[[1]]
medianPosterior <- median(delta[delta >= 0])
if (any(is.na(c(CIlow, CIhigh, medianPosterior)))) {
CIlow <- .qShiftedT(0.025, parameters, oneSided = "right")
CIhigh <- .qShiftedT(0.975, parameters, oneSided = "right")
medianPosterior <- .qShiftedT(0.5, parameters, oneSided = "right")
}
}
if (oneSided == "left") {
CIlow <- quantile(delta[delta <= 0], probs = 0.025)[[1]]
CIhigh <- quantile(delta[delta <= 0], probs = 0.975)[[1]]
medianPosterior <- median(delta[delta <= 0])
if (any(is.na(c(CIlow, CIhigh, medianPosterior)))) {
CIlow <- .qShiftedT(0.025, parameters, oneSided = "left")
CIhigh <- .qShiftedT(0.975, parameters, oneSided = "left")
medianPosterior <- .qShiftedT(0.5, parameters, oneSided = "left")
}
}
posteriorLine <- .dposteriorShiftedT(x = seq(min(xticks), max(xticks),
length.out = 1000), parameters = parameters, oneSided = oneSided)
xlim <- c(min(CIlow, range(xticks)[1]), max(range(xticks)[2], CIhigh)) plot(1, 1, xlim = xlim, ylim = range(yticks), ylab = "", xlab = "", type = "n", axes = FALSE) lines(seq(min(xticks), max(xticks), length.out = 1000), posteriorLine, lwd = lwd) lines(seq(min(xticks), max(xticks), length.out = 1000), .dprior(seq(min(xticks), max(xticks), length.out = 1000), r = r, oneSided = oneSided), lwd = lwd, lty = 3) axis(1, at = xticks, labels = xlabels, cex.axis = cexAxis, lwd = lwdAxis) axis(2, at = yticks, labels = ylabels, , cex.axis = cexAxis, lwd = lwdAxis) if (nchar(ylabels[length(ylabels)]) > 4) {
mtext(text = "Density", side = 2, las = 0, cex = cexYlab, line = 4)
} else if (nchar(ylabels[length(ylabels)]) == 4) {
mtext(text = "Density", side = 2, las = 0, cex = cexYlab, line = 3.25)
} else if (nchar(ylabels[length(ylabels)]) < 4) {
mtext(text = "Density", side = 2, las = 0, cex = cexYlab, line = 2.85)
}
mtext(expression(paste("Effect size", ~delta)), side = 1, cex = cexXlab,
line = 2.5)
points(0, .dprior(0, r, oneSided = oneSided), col = "black", pch = 21,
bg = "grey", cex = cexPoints)
if (oneSided == FALSE) {
heightPosteriorAtZero <- .dposteriorShiftedT(0, parameters = parameters,
oneSided = oneSided)
} else if (oneSided == "right") {
posteriorLineLargerZero <- posteriorLine[posteriorLine > 0]
heightPosteriorAtZero <- posteriorLineLargerZero[1]
} else if (oneSided == "left") {
posteriorLineLargerZero <- posteriorLine[posteriorLine > 0]
heightPosteriorAtZero <- posteriorLineLargerZero[length(posteriorLineLargerZero)]
}
points(0, heightPosteriorAtZero, col = "black", pch = 21, bg = "grey",
cex = cexPoints)
### 95% credible interval
# enable plotting in margin
par(xpd = TRUE)
yCI <- grconvertY(dmax, "user", "ndc") + 0.04
yCI <- grconvertY(yCI, "ndc", "user")
arrows(CIlow, yCI, CIhigh, yCI, angle = 90, code = 3, length = 0.1,
lwd = lwd)
medianText <- formatC(medianPosterior, digits = 3, format = "f")
if (addInformation) {
# display BF10 value
offsetTopPart <- 0.06
yy <- grconvertY(0.75 + offsetTopPart, "ndc", "user")
yy2 <- grconvertY(0.806 + offsetTopPart, "ndc", "user")
xx <- min(xticks) if (BF10 >= 1000000 | BF01 >= 1000000) {
BF10t <- formatC(BF10, 3, format = "e")
BF01t <- formatC(BF01, 3, format = "e")
}
if (BF10 < 1000000 & BF01 < 1000000) {
BF10t <- formatC(BF10, 3, format = "f")
BF01t <- formatC(BF01, 3, format = "f")
}
if (oneSided == FALSE) {
text(xx, yy2, bquote(BF[10] == .(BF10t)), cex = cexTextBF,
pos = 4)
text(xx, yy, bquote(BF[0][1] == .(BF01t)), cex = cexTextBF,
pos = 4)
}
if (oneSided == "right") {
text(xx, yy2, bquote(BF["+"][0] == .(BF10t)), cex = cexTextBF,
pos = 4)
text(xx, yy, bquote(BF[0]["+"] == .(BF01t)), cex = cexTextBF,
pos = 4)
}
if (oneSided == "left") {
text(xx, yy2, bquote(BF["-"][0] == .(BF10t)), cex = cexTextBF,
pos = 4)
text(xx, yy, bquote(BF[0]["-"] == .(BF01t)), cex = cexTextBF,
pos = 4)
}
yy <- grconvertY(0.756 + offsetTopPart, "ndc", "user")
yy2 <- grconvertY(0.812 + offsetTopPart, "ndc", "user")
CIText <- paste("95% CI: [", bquote(.(formatC(CIlow, 3, format = "f"))),
", ", bquote(.(formatC(CIhigh, 3, format = "f"))), "]", sep = "")
medianLegendText <- paste("median =", medianText)
text(max(xticks), yy2, medianLegendText, cex = 1.1, pos = 2)
text(max(xticks), yy, CIText, cex = 1.1, pos = 2)
# probability wheel
if (max(nchar(BF10t), nchar(BF01t)) <= 4) {
xx <- grconvertX(0.44, "ndc", "user")
}
if (max(nchar(BF10t), nchar(BF01t)) == 5) {
xx <- grconvertX(0.44 + 0.001 * 5, "ndc", "user")
}
if (max(nchar(BF10t), nchar(BF01t)) == 6) {
xx <- grconvertX(0.44 + 0.001 * 6, "ndc", "user")
}
if (max(nchar(BF10t), nchar(BF01t)) == 7) {
xx <- grconvertX(0.44 + 0.002 * max(nchar(BF10t), nchar(BF01t)),
"ndc", "user")
}
if (max(nchar(BF10t), nchar(BF01t)) == 8) {
xx <- grconvertX(0.44 + 0.003 * max(nchar(BF10t), nchar(BF01t)), "ndc", "user") } if (max(nchar(BF10t), nchar(BF01t)) > 8) {
xx <- grconvertX(0.44 + 0.005 * max(nchar(BF10t), nchar(BF01t)),
"ndc", "user")
}
yy <- grconvertY(0.788 + offsetTopPart, "ndc", "user")
# make sure that colored area is centered
radius <- 0.06 * diff(range(xticks))
A <- radius^2 * pi
alpha <- 2/(BF01 + 1) * A/radius^2
startpos <- pi/2 - alpha/2
# draw probability wheel
plotrix::floating.pie(xx, yy, c(BF10, 1), radius = radius, col = c("darkred",
"white"), lwd = 2, startpos = startpos)
yy <- grconvertY(0.865 + offsetTopPart, "ndc", "user")
yy2 <- grconvertY(0.708 + offsetTopPart, "ndc", "user")
if (oneSided == FALSE) {
text(xx, yy, "data|H1", cex = cexCI)
text(xx, yy2, "data|H0", cex = cexCI)
}
if (oneSided == "right") {
text(xx, yy, "data|H+", cex = cexCI)
text(xx, yy2, "data|H0", cex = cexCI)
}
if (oneSided == "left") {
text(xx, yy, "data|H-", cex = cexCI)
text(xx, yy2, "data|H0", cex = cexCI)
}
# add legend
CIText <- paste("95% CI: [", bquote(.(formatC(CIlow, 3, format = "f"))),
" ; ", bquote(.(formatC(CIhigh, 3, format = "f"))), "]", sep = "")
medianLegendText <- paste("median =", medianText)
}
mostPosterior <- mean(delta > mean(range(xticks)))
if (mostPosterior >= 0.5) {
legendPosition <- min(xticks)
legend(legendPosition, max(yticks), legend = c("Posterior", "Prior"),
lty = c(1, 3), bty = "n", lwd = c(lwd, lwd), cex = cexLegend,
xjust = 0, yjust = 1, x.intersp = 0.6, seg.len = 1.2)
} else {
legendPosition <- max(xticks)
legend(legendPosition, max(yticks), legend = c("Posterior", "Prior"),
lty = c(1, 3), bty = "n", lwd = c(lwd, lwd), cex = cexLegend,
xjust = 1, yjust = 1, x.intersp = 0.6, seg.len = 1.2)
}
}
### generate data ###
set.seed(1)
x <- rnorm(30, 0.15)
### calculate Bayes factor ###
library(BayesFactor)
BF <- extractBF(ttestBF(x, rscale = "medium"), onlybf = TRUE)
### plot ###
.plotPosterior.ttest(x = x, rscale = "medium", BF = BF)
#monte carlo t test simulation
#install.packages("MonteCarlo")
library(MonteCarlo)
# Define function that generates data and applies the method of interest
ttest<-function(n,loc,scale){
# generate sample:
sample<-rnorm(n, loc, scale)
# calculate test statistic:
stat<-sqrt(n)*mean(sample)/sd(sample)
# get test decision:
decision<-abs(stat)>1.96
# return result:
return(list("decision"=decision))
}
# define parameter grid:
n_grid<-c(50,100,250,500)
loc_grid<-seq(0,1,0.2)
scale_grid<-c(1,2)
# collect parameter grids in list:
param_list=list("n"=n_grid, "loc"=loc_grid, "scale"=scale_grid)
# run simulation:
MC_result<-MonteCarlo(func=ttest, nrep=1000, param_list=param_list)
summary(MC_result)
#install.packages("RWordPress", repos="http://www.omegahat.org/R", build=TRUE)
library(RWordPress)
options(WordPressLogin=c(user="password"),
WordPressURL="http://your_wp_installation.org/xmlrpc.php")
#[code lang='r']
#...
#[/code]
knit_hooks$set(output=function(x, options) paste("\\[code\\]\n", x, "\\[/code\\]\n", sep=""))
knit_hooks$set(source=function(x, options) paste("\\[code lang='r'\\]\n", x, "\\[/code\\]\n", sep=""))
knit2wp <- function(file) {
require(XML)
content <- readLines(file)
content <- htmlTreeParse(content, trim=FALSE)
## WP will add the h1 header later based on the title, so delete here
content$children$html$children$body$children$h1 <- NULL
content <- paste(capture.output(print(content$children$html$children$body,
indent=FALSE, tagSeparator="")),
collapse="\n")
content <- gsub("<?.body>", "", content) # remove body tag
## enclose code snippets in SyntaxHighlighter format
content <- gsub("<?pre><code class=\"r\">", "\\[code lang='r'\\]\\\n",
content)
content <- gsub("<?pre><code class=\"no-highlight\">", "\\[code\\]\\\n",
content)
content <- gsub("<?pre><code>", "\\[code\\]\\\n", content)
content <- gsub("<?/code></pre>", "\\[/code\\]\\\n", content)
return(content)
}
newPost(content=list(description=knit2wp('rerWorkflow.html'),
title='Workflow: Post R markdown to WordPress',
categories=c('R')),
publish=FALSE)
postID <- 99 # post id returned by newPost()
editPost(postID,
content=list(description=knit2wp('rerWorkflow.html'),
title='Workflow: Post R markdown to WordPress',
categories=c('R')),
publish=FALSE)